bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7021_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
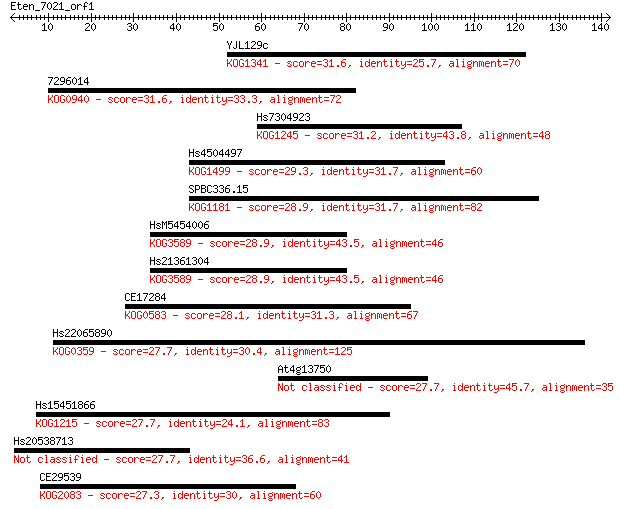
YJL129c 31.6 0.45
7296014 31.6 0.50
Hs7304923 31.2 0.64
Hs4504497 29.3 2.4
SPBC336.15 28.9 3.2
HsM5454006 28.9 3.3
Hs21361304 28.9 3.4
CE17284 28.1 6.2
Hs22065890 27.7 7.2
At4g13750 27.7 7.2
Hs15451866 27.7 7.8
Hs20538713 27.7 8.2
CE29539 27.3 8.5
> YJL129c
Length=1235
Score = 31.6 bits (70), Expect = 0.45, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 52 GEVSSRSSLGSSSGNSNSSGNSTCTSSPAPCHSPFSIPGTAGAPAAVPAADGIAKDTKQL 111
GE +S + +SN+ + + P P F P + PA VP ++ AK
Sbjct 246 GEYQENNSYSTVGSSSNTVADESLNQKPKPSSLRFDEPHSKQRPARVP-SEKFAKRRGSR 304
Query 112 NTDPSEVHRS 121
+ P++++RS
Sbjct 305 DISPADMYRS 314
> 7296014
Length=949
Score = 31.6 bits (70), Expect = 0.50, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 11/72 (15%)
Query 10 EMRGVAVTQVGSHKKELTSPVTVASSSASASCGLGRLAELAEGEVSSRSSLGSSSGNSNS 69
+++ VA Q G+ + +P V+ ++A SC + G V +R L SSSGNSN
Sbjct 183 QIQPVAGQQNGNPPVQAVNPSVVSDAAAGRSC-------MIYGGVRARMRLRSSSGNSNG 235
Query 70 SGNSTCTSSPAP 81
T SP P
Sbjct 236 GE----TRSPLP 243
> Hs7304923
Length=1972
Score = 31.2 bits (69), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 59 SLGSSSGNSNSSGNSTCTSSPAPCHSPFSIPGTAGAPAAVPAADGIAK 106
SLGS G S +GNS TS+ A S +P A +A PAA +AK
Sbjct 1457 SLGSGLGLSEGNGNSFLTSNVASSKSESPVPQNEKATSAQPAAVEVAK 1504
> Hs4504497
Length=360
Score = 29.3 bits (64), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query 43 LGRLAELAEGEVSSRSSLGSSSGNSNSSGNSTCTSSPAPCHSPFSIPGTAGAPAAVPAAD 102
L + + +G+V +S++SG +T SS +PC +P S+PGT+ AP + D
Sbjct 125 LDHVVTIIKGKVEEVELPVEKVASSSASGWAT-ASSTSPCSTPCSMPGTSVAPDGLIFPD 183
> SPBC336.15
Length=910
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 40/83 (48%), Gaps = 1/83 (1%)
Query 43 LGRLAELAEGEVSSRSSLGSSSGNSNSSGNSTCTSSPAPCHSPFSIPGTAGAPAAVPAAD 102
G+ EL+ E SR + +S NS++ + T S P P +IP + + P +
Sbjct 637 YGKERELSNNEFPSRQTKTVTSANSSNIRDMEHTISDKPRSEPDAIPSSKSMHSNKPFEE 696
Query 103 GIAK-DTKQLNTDPSEVHRSRRS 124
K TK+L T+PS V+ S S
Sbjct 697 KSEKPTTKRLVTNPSNVNASWHS 719
> HsM5454006
Length=441
Score = 28.9 bits (63), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 34 SSSASASCGLGRLAELAEGEVSSRSSLGSSSGNSNSS-GNSTCTSSP 79
S ++SAS LG LA ++ S R SLGS+ G S S G C P
Sbjct 110 SLNSSASLDLGFLAFVSSKSESHRKSLGSTEGESESRPGKYCCVYLP 156
> Hs21361304
Length=566
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 34 SSSASASCGLGRLAELAEGEVSSRSSLGSSSGNSNSS-GNSTCTSSP 79
S ++SAS LG LA ++ S R SLGS+ G S S G C P
Sbjct 263 SLNSSASLDLGFLAFVSSKSESHRKSLGSTEGESESRPGKYCCVYLP 309
> CE17284
Length=726
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 8/67 (11%)
Query 28 SPVTVASSSASASCGLGRLAELAEGEVSSRSSLGSSSGNSNSSGNSTCTSSPAPCHSPFS 87
SP++V SS +++ LG A + G V +S+G + G STC S P
Sbjct 444 SPMSVRSSDSAS---LGSAATPSRGGVKDNDKENASTGKNYRMGASTCKS-----RGPLK 495
Query 88 IPGTAGA 94
I G A
Sbjct 496 ITGVGKA 502
> Hs22065890
Length=419
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 57/139 (41%), Gaps = 22/139 (15%)
Query 11 MRGVAVTQVGSHKKELTSPVTVASSSASASCGLGRLAELAEGEV---SSRSSLGSSSGNS 67
+R +AV ++G K+L V V+S + G + + V S LGS G +
Sbjct 68 LRQLAVEKLGGATKQL---VAVSSQKEKSK---GEIVQAQPDYVKPSSVTVMLGSLQGLN 121
Query 68 NSSGNSTCTSSPAPCHSPFSIPGTAGAPAAVPAADGIAKDTKQLNTDPSEVH-------- 119
N + NS T+S P PF P + + GI + TK DP +H
Sbjct 122 NCAINSPMTASKVPVCLPFLKPSVLCI-ISREGSLGIFR-TKFQRQDPVTLHLASANQKS 179
Query 120 ---RSRRSVDDDDPQVNRG 135
R+ D+ PQ+ RG
Sbjct 180 YLTRNPIHWLDERPQIPRG 198
> At4g13750
Length=2137
Score = 27.7 bits (60), Expect = 7.2, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query 64 SGNSNSSGNSTCTSSPAPCHSPFSIPG-TAGAPAAV 98
SG N +G S+ S P P HS I G T+G AA+
Sbjct 1940 SGYDNCAGTSSRASEPNPLHSMHMISGSTSGNQAAM 1975
> Hs15451866
Length=700
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 30/83 (36%), Gaps = 0/83 (0%)
Query 7 GPEEMRGVAVTQVGSHKKELTSPVTVASSSASASCGLGRLAELAEGEVSSRSSLGSSSGN 66
G +E QV +K P + AS C + E E +SLG+ G+
Sbjct 114 GSDESEATCTKQVCPAEKLSCGPTSHKCVPASWRCDGEKDCEGGADEAGCATSLGTCRGD 173
Query 67 SNSSGNSTCTSSPAPCHSPFSIP 89
G+ TC + C+ P
Sbjct 174 EFQCGDGTCVLAIKHCNQEQDCP 196
> Hs20538713
Length=426
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 2 RRHAHGPEEMRGVAVTQVGSHKKELTSPVTVASSSASASCG 42
RR HG +E R V + G HKK + + + A S S G
Sbjct 340 RRLFHGTKEYRHQRVARAGKHKKTVCTRTSYADSCQVLSLG 380
> CE29539
Length=838
Score = 27.3 bits (59), Expect = 8.5, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query 8 PEEMRGVAVTQVGSHKKELTSPVTVASSSASASCGLGRLAELAEGEVSSRSSLGSSSGNS 67
P M G+ + +GS K + T+P + A ++C G A L E +R+S +S+G++
Sbjct 154 PAAMCGLTILNIGS-KLQNTAPRGALIAIAVSACFYGAAAMLDYVEFFARTSTSNSTGSA 212
Lambda K H
0.304 0.119 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40