bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6634_orf3
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
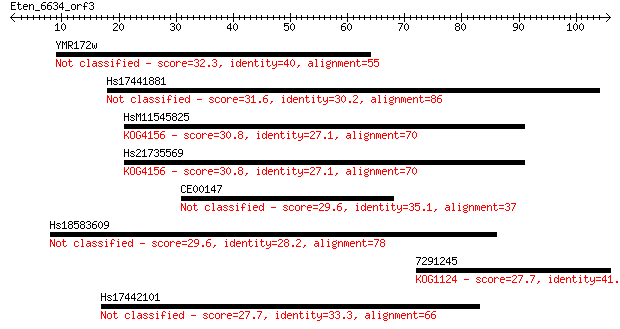
YMR172w 32.3 0.19
Hs17441881 31.6 0.33
HsM11545825 30.8 0.67
Hs21735569 30.8 0.70
CE00147 29.6 1.4
Hs18583609 29.6 1.5
7291245 27.7 5.4
Hs17442101 27.7 5.7
> YMR172w
Length=719
Score = 32.3 bits (72), Expect = 0.19, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 13/68 (19%)
Query 9 RHASICRRRGSLHSKLDNSCSG-------AATEPEATSE------PAATQEMALYPGAAT 55
++ASI +RR SLH K NS +G +AT+P S+ A T ++ T
Sbjct 576 KNASINKRRRSLHHKKSNSLNGRRKLHGESATKPNINSDLHYRILKAPTDVKTIWEEYDT 635
Query 56 ATRGRPCV 63
RG+P +
Sbjct 636 GIRGKPSI 643
> Hs17441881
Length=394
Score = 31.6 bits (70), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 36/94 (38%), Gaps = 8/94 (8%)
Query 18 GSLHSKLDNSCSGAATEPEATSEPAATQEMALYP-GAATATRGRPCVGARRQQQQREQRQ 76
G L LD+ G E EP T+ A YP G G+R +Q++ E R
Sbjct 249 GPLVRILDSGDLGLFDEYSGQGEPDQTEAPACYPRGHEVVNSSLDSAGSRTKQEETEGRL 308
Query 77 NFCCPCPQRQSLLRFA-------ALCVVCILCFC 103
C P R + L F ++CV I+
Sbjct 309 VSCWPVATRLTELEFKPMYGEPNSVCVWPIMLLV 342
> HsM11545825
Length=1332
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query 21 HSKLDNSCSGAATEPEATSEPAATQEMALYPGAATATRGRPCVGARRQQQQREQRQNFCC 80
HSK +GA E E + P ++E + G+ + R + V + Q +Q+Q+
Sbjct 361 HSKGSEQTTGAENEVETNALPVVSKETQIITGSDESCR-KDLVKNEELEIQEKQKQSDIR 419
Query 81 PCPQRQSLLR 90
P P S+L+
Sbjct 420 PSPGDSSVLQ 429
> Hs21735569
Length=1339
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query 21 HSKLDNSCSGAATEPEATSEPAATQEMALYPGAATATRGRPCVGARRQQQQREQRQNFCC 80
HSK +GA E E + P ++E + G+ + R + V + Q +Q+Q+
Sbjct 361 HSKGSEQTTGAENEVETNALPVVSKETQIITGSDESCR-KDLVKNEELEIQEKQKQSDIR 419
Query 81 PCPQRQSLLR 90
P P S+L+
Sbjct 420 PSPGDSSVLQ 429
> CE00147
Length=405
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 31 AATEPEATSEPAATQEMALYPGAATATRGRPCVGARR 67
+ T P AT+ A+ + ++ PG + +T G P VG R+
Sbjct 179 SKTPPIATTRFASLRHLSSAPGTSASTSGSPSVGIRK 215
> Hs18583609
Length=565
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 38/86 (44%), Gaps = 12/86 (13%)
Query 8 TRHASICRRRGSLHSKLDNSCSGAATEPEATSEPAATQEMALYPGAATATRGRPCVGARR 67
TR+ + +GS + L+N+ + P+ + P T E+A +G G R+
Sbjct 34 TRNPELGEEKGSSQAYLENAAFKGCSAPKKSMPPTTTLEVA----KERDEQGDSPKGWRQ 89
Query 68 QQQQ--------REQRQNFCCPCPQR 85
++Q+ R+Q QNF PQ
Sbjct 90 ERQRNMYPGVSLRDQDQNFISDSPQN 115
> 7291245
Length=870
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query 72 REQRQNFCCPCPQRQSLLRFAALCVVCILCFCRS 105
RE+ + PQR +LL F +CV CI+C+ S
Sbjct 19 REREREGATNSPQR-NLLEFLCICVACIVCYYNS 51
> Hs17442101
Length=425
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 8/70 (11%)
Query 17 RGSLHSKLDNSCSGAATEPEATSEPAATQEMAL----YPGAATATRGRPCVGARRQQQQR 72
R SLHS L S + + E EAT ++ EM + Y + +RG+ +RRQ
Sbjct 67 RASLHSTLGLSWNNSMKEAEATLLQGSSFEMQVLCGKYIDKKSLSRGK----SRRQWFHV 122
Query 73 EQRQNFCCPC 82
E + C C
Sbjct 123 ELGKRLTCVC 132
Lambda K H
0.324 0.131 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1170944580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40