bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6634_orf2
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
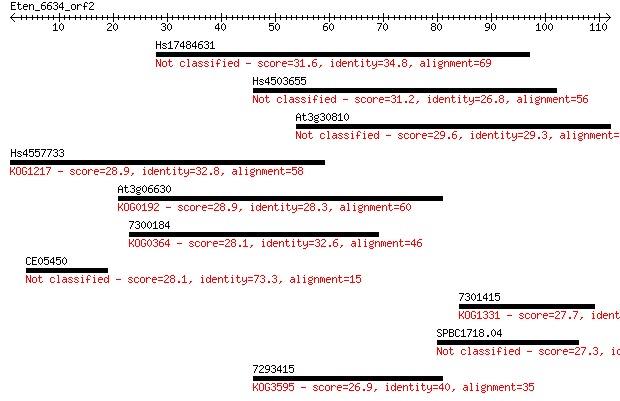
Hs17484631 31.6 0.40
Hs4503655 31.2 0.53
At3g30810 29.6 1.6
Hs4557733 28.9 2.2
At3g06630 28.9 2.7
7300184 28.1 3.9
CE05450 28.1 4.0
7301415 27.7 5.1
SPBC1718.04 27.3 7.6
7293415 26.9 8.3
> Hs17484631
Length=699
Score = 31.6 bits (70), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 29/72 (40%), Gaps = 12/72 (16%)
Query 28 QQAGQLMLRSSNRARSNLRASSHAGNGALSRGSDSNTGETLRRCQ---EAAATTGAKAKL 84
+Q RSS + S L +SH G L RG RCQ E A A +
Sbjct 411 KQGFNFANRSSEKCASQLPTASHVGKTYLGRGV---------RCQGGWETARKPNAPREA 461
Query 85 LLPLSSTPVPAT 96
L P+S P P T
Sbjct 462 LFPISWWPFPPT 473
> Hs4503655
Length=1455
Score = 31.2 bits (69), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 24/56 (42%), Gaps = 0/56 (0%)
Query 46 RASSHAGNGALSRGSDSNTGETLRRCQEAAATTGAKAKLLLPLSSTPVPATFCCTL 101
R+ H+ N L G + + + R QE A + L++PL TP F +
Sbjct 996 RSYDHSENSDLVFGGRTGNEDIISRLQEMVADLELQQDLIVPLGHTPSQEHFLFEI 1051
> At3g30810
Length=791
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 27/61 (44%), Gaps = 8/61 (13%)
Query 54 GALSRGSDSNTGETLRRCQEAAATTGAKAKLLLP---LSSTPVPATFCCTLCCLYSLLLP 110
G +R ++S+ + RRCQ T ++LP L VP +CC + +P
Sbjct 65 GGGTRMTNSDLVDLRRRCQVPVGVT-----MILPRPPLHRENVPVGWCCAYANFFEKYMP 119
Query 111 E 111
E
Sbjct 120 E 120
> Hs4557733
Length=1821
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 1 TRTQDLTCDTSRRDSACLHLPPPGQLAQQAGQLMLRSSNRARSNLRASSHAGNGALSR 58
+R Q C + R + C + P + Q +L R NLR SS AG G L+R
Sbjct 202 SRPQLCVCRSGFRGARCEEVIPDEEFDPQNSRLAPRRWAERSPNLRRSSAAGEGTLAR 259
> At3g06630
Length=671
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 21 PPPGQLAQQAGQLMLRSSNRARSNLRASSHAGNGALSRGSDSNTGETLRRCQEAAATTGA 80
P +A + L + SN+ RS +RA ++ G G+TL ++ AA++GA
Sbjct 216 PIQVSIASKISSLASKLSNKVRSKMRAGDNSACGDSHHSDHDVFGDTLSDHRDDAASSGA 275
> 7300184
Length=544
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 23 PGQLAQQAGQLMLRSSNRARSNLRASSHAGNGALSRGSDSNTGETL 68
P LAQ G +R+ R+ + +SH G+G + G D +GE +
Sbjct 444 PRTLAQNCGANTIRALTALRA--KHASHTGDGVCAWGIDGESGEIV 487
> CE05450
Length=721
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 11/15 (73%), Positives = 13/15 (86%), Gaps = 0/15 (0%)
Query 4 QDLTCDTSRRDSACL 18
QDL+CD S+R SACL
Sbjct 416 QDLSCDFSKRASACL 430
> 7301415
Length=1270
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 84 LLLPLSSTPVPATFCCTLCCLYSLL 108
L+LP SST P T+C ++ C S L
Sbjct 569 LILPASSTSAPVTYCSSVECTMSNL 593
> SPBC1718.04
Length=675
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 80 AKAKLLLPLSSTPVPATFCCTLCCLY 105
A KLL+ L TP+ +F LCC Y
Sbjct 444 ATWKLLVALGMTPILYSFYALLCCYY 469
> 7293415
Length=4081
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 46 RASSHAGNGALSRGSDSNTGETLRRCQEAAATTGA 80
RAS H N + S+ + S TG R TTGA
Sbjct 118 RASKHRLNFSTSKSAGSGTGNKSRSASTTKITTGA 152
Lambda K H
0.317 0.127 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40