bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6517_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
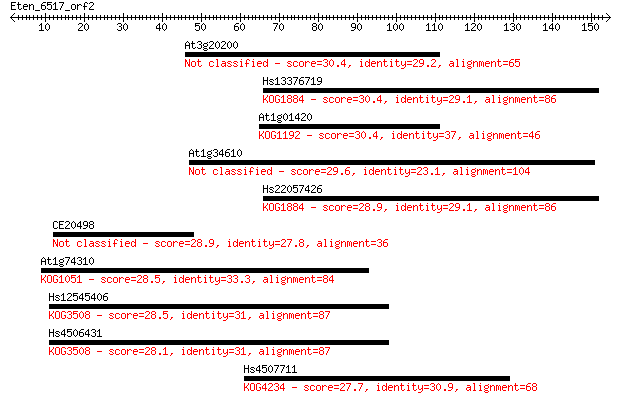
At3g20200 30.4 1.3
Hs13376719 30.4 1.6
At1g01420 30.4 1.6
At1g34610 29.6 2.6
Hs22057426 28.9 4.2
CE20498 28.9 4.3
At1g74310 28.5 5.3
Hs12545406 28.5 5.5
Hs4506431 28.1 7.3
Hs4507711 27.7 9.7
> At3g20200
Length=750
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 35/70 (50%), Gaps = 5/70 (7%)
Query 46 SQHRGARLRRLSSDDEGACEQGVVAEE-----VAEEVAEEVAKEVEGAEGGVSNEEAQRA 100
++ L+R ++EG ++G ++E+ V +E A + AK+ G ++ E QR
Sbjct 334 AKQEAKELQRQKIEEEGWVQEGQLSEKSTKSIVEKERAHKAAKDASETAGKIAELETQRR 393
Query 101 REEAVQAFRD 110
EA +F D
Sbjct 394 AIEAAGSFSD 403
> Hs13376719
Length=425
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 14/89 (15%)
Query 66 QGVVAEEVAEEVAEEVAKEVEGAEGGVSNEEAQRAREEAVQAFRDYITA---RTAAALRA 122
QG+ + VAE VAEEV +E+ + G +++ EE+ Q F T RT ++
Sbjct 24 QGLKPDTVAELVAEEVTRELLTSSQGTGHKQMFNPTEES-QTFLQLTTLCQDRTLVGMKL 82
Query 123 AEFLATAARLTHQCGIVPHDEDEQAEQLL 151
+ +++ VPH E +LL
Sbjct 83 LDKISS----------VPHGELSCTTELL 101
> At1g01420
Length=481
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 65 EQGVVAEEVAEEVAEEVAKEVEGAEGGVSNEEAQRAREEAVQAFRD 110
E GVV E EVA V +EG EG ++ + +E +V+ RD
Sbjct 409 EDGVVGRE---EVARVVKGLIEGEEGNAVRKKMKELKEGSVRVLRD 451
> At1g34610
Length=997
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 24/104 (23%), Positives = 43/104 (41%), Gaps = 1/104 (0%)
Query 47 QHRGARLRRLSSDDEGACEQGVVAEEVAEEVAEEVAKEVEGAEGGVSNEEAQRAREEAVQ 106
Q R + ++D G+ G+ E V +EVA+E K V + ++E E V+
Sbjct 425 QQEVPRAGEMENNDSGSAGHGL-REAVIKEVADEFEKRVLKIQQSNNDELKSYFEAELVK 483
Query 107 AFRDYITARTAAALRAAEFLATAARLTHQCGIVPHDEDEQAEQL 150
++ + T + A L A + +C D + EQ+
Sbjct 484 QKKELVAILTESVREAVSGLNRQAGIVGECSKCVADAHSKQEQI 527
> Hs22057426
Length=2407
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 37/86 (43%), Gaps = 8/86 (9%)
Query 66 QGVVAEEVAEEVAEEVAKEVEGAEGGVSNEEAQRAREEAVQAFRDYITARTAAALRAAEF 125
QG+ + VAE VAEEV +E+ + G +++ EE+ Q F T L
Sbjct 2042 QGLKPDTVAELVAEEVTRELLTSSQGTGHKQMFNPTEES-QTFLQLTTLCQDRTL----- 2095
Query 126 LATAARLTHQCGIVPHDEDEQAEQLL 151
+L + VPH E +LL
Sbjct 2096 --VGMKLLDKISSVPHGELSCTTELL 2119
> CE20498
Length=348
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 10/36 (27%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 12 RRSRGARVLLATSALAATLAVAFLVMHCFIVLKNSQ 47
++ G +L ++ + T+ +AF ++H F+ L+ SQ
Sbjct 208 KKITGWSILAYSTNIIVTIEIAFFIIHSFLCLRKSQ 243
> At1g74310
Length=911
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 35/84 (41%), Gaps = 3/84 (3%)
Query 9 ILRRRSRGARVLLATSALAATLAVAFLVMHCFIVLKNSQHRGARLRRLSSDDEGACEQGV 68
IL RR++ VL+ + T V L ++K RL S D GA G
Sbjct 194 ILSRRTKNNPVLIGEPGVGKTAVVEGLAQR---IVKGDVPNSLTDVRLISLDMGALVAGA 250
Query 69 VAEEVAEEVAEEVAKEVEGAEGGV 92
EE + V KEVE AEG V
Sbjct 251 KYRGEFEERLKSVLKEVEDAEGKV 274
> Hs12545406
Length=870
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 41/90 (45%), Gaps = 12/90 (13%)
Query 11 RRRSRGARVLLATSALAATLAVAFLVMHCFIVLKNSQHRGARLRRLSSDDEGACEQGVVA 70
RRR R +L + + T ++FL FIV + + L +D EQG++
Sbjct 104 RRR---VRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTD-----EQGLIV 155
Query 71 EEVAEEVAEEVAKEVEGA---EGGVSNEEA 97
E++ EEV E EG G +S +EA
Sbjct 156 EDLVEEVGREEDPH-EGKIWFHGKISKQEA 184
> Hs4506431
Length=1047
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 41/90 (45%), Gaps = 12/90 (13%)
Query 11 RRRSRGARVLLATSALAATLAVAFLVMHCFIVLKNSQHRGARLRRLSSDDEGACEQGVVA 70
RRR R +L + + T ++FL FIV + + L +D EQG++
Sbjct 281 RRR---VRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTD-----EQGLIV 332
Query 71 EEVAEEVAEEVAKEVEGA---EGGVSNEEA 97
E++ EEV E EG G +S +EA
Sbjct 333 EDLVEEVGREEDPH-EGKIWFHGKISKQEA 361
> Hs4507711
Length=292
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 61 EGACEQGVVAEEVAEEVAEEVAKEVEGAEGGVSNEEAQRAREEAVQAFR--DYITARTAA 118
E + V + E+ EE E+ K + E EE+ R +EE + F+ DYI A ++
Sbjct 80 ENKSNEDVNSSELDEEYLIELEKNMSDEEKQKRREESTRLKEEGNEQFKKGDYIEAESSY 139
Query 119 ALRAAEFLAT 128
+ RA E +
Sbjct 140 S-RALEMCPS 148
Lambda K H
0.317 0.128 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40