bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5237_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
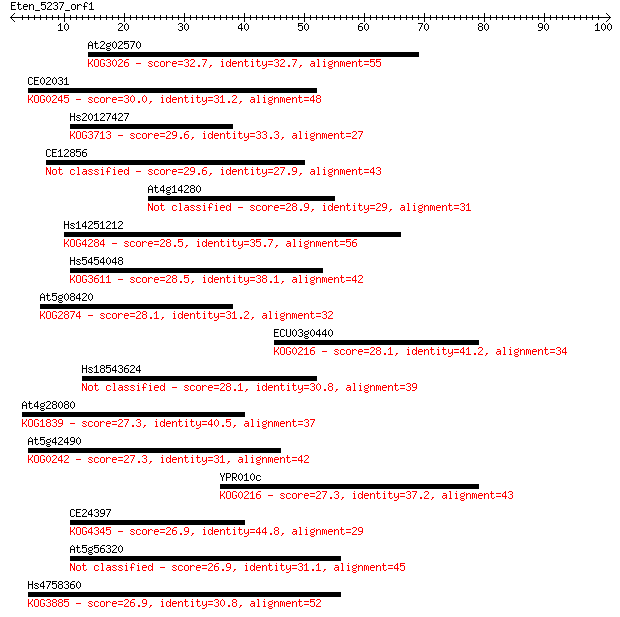
At2g02570 32.7 0.17
CE02031 30.0 1.1
Hs20127427 29.6 1.3
CE12856 29.6 1.6
At4g14280 28.9 2.7
Hs14251212 28.5 3.0
Hs5454048 28.5 3.0
At5g08420 28.1 4.1
ECU03g0440 28.1 4.1
Hs18543624 28.1 4.5
At4g28080 27.3 6.3
At5g42490 27.3 6.9
YPR010c 27.3 7.6
CE24397 26.9 8.5
At5g56320 26.9 8.5
Hs4758360 26.9 8.6
> At2g02570
Length=316
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 26/55 (47%), Gaps = 6/55 (10%)
Query 14 QKKQQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRRSKMRIPYGSFAAVALVGEG 68
Q K+Q +WQQ Q + + K+G GRK+ S + P F V + G G
Sbjct 243 QNKKQNDWQQF------QTTKAKTKKVGFFTGRKKESIFKSPEDPFGKVGVTGSG 291
> CE02031
Length=928
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Query 4 LRLQLEPNQYQKKQQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRRSK 51
LR QL NQ K+ + +++WQQK ++++K++S K+ + + K
Sbjct 403 LRRQLAENQ---KEMEEMEKSWQQKIAEEAAKHASGASEKVEMEAKKK 447
> Hs20127427
Length=491
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 11 NQYQKKQQQNWQQNWQQKQQQDSSKNS 37
N+YQ+++++N +++W QK S+ +S
Sbjct 119 NRYQERKEENHEKDWDQKSHDVSTDSS 145
> CE12856
Length=592
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 12/45 (26%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 7 QLEPNQYQKK--QQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRR 49
++E N +++ ++ +NWQ+++QQD +S I S + K +
Sbjct 256 EIEENSFEEDVVPEEKNMKNWQERRQQDGEYDSVSISSSLVNKNK 300
> At4g14280
Length=848
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 9/31 (29%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 24 NWQQKQQQDSSKNSSKIGSKIGRKRRSKMRI 54
NW+ K Q+D ++++I S++ K+++ +R+
Sbjct 361 NWRDKNQEDVRMSAAEILSRLASKKQNSLRV 391
> Hs14251212
Length=824
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 4/60 (6%)
Query 10 PNQYQKKQQQNWQQNWQ-QKQQQD---SSKNSSKIGSKIGRKRRSKMRIPYGSFAAVALV 65
PNQ KKQ Q ++ Q QK D SS+N+S G + K +K ++P S + ++
Sbjct 468 PNQPLKKQIQKIERTLQIQKAHGDHMASSRNNSVSGLSVKSKNNTKQKLPVKSHSECGII 527
> Hs5454048
Length=751
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 11 NQYQKKQQQNWQQ-NWQQKQQQDSSKNSSKIGSKIGRKRRSKM 52
NQY K +Q QQ + QK + D K + I S+ R RR+++
Sbjct 706 NQYCKDTRQQHQQGDESQKMRGDYGKLKALINSRKSRNRRNQL 748
> At5g08420
Length=391
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 10/32 (31%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 6 LQLEPNQYQKKQQQNWQQNWQQKQQQDSSKNS 37
+QLE +Y ++ ++ WQ+KQ++ S K++
Sbjct 278 MQLESGEYFMSDKKKSEKKWQEKQEKQSEKST 309
> ECU03g0440
Length=1062
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query 45 GRKRRSKMRIPYGSFAAVALVGEGIIYLRRDRLV 78
GRK+R +R +G AL+ G YL +DRL+
Sbjct 972 GRKKRGGIR--FGEMERDALISHGASYLLQDRLL 1003
> Hs18543624
Length=1343
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 13 YQKKQQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRRSK 51
Y Q +W+Q QK + S NS+++ + G++ R+K
Sbjct 940 YADSSQLSWEQRALQKPRTPSHTNSAQLTQRGGKEIRTK 978
> At4g28080
Length=1791
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 3 LLRLQLEPNQYQKKQQQNWQQNW-QQKQQQDSSKNSSK 39
L +L++EP++Y K + W Q Q Q SSK+ SK
Sbjct 523 LKKLEIEPSRYSKPIRWELGACWVQHLQNQASSKSESK 560
> At5g42490
Length=1087
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 4 LRLQLEPNQYQKKQQQNWQQNWQQKQQQDSSK--NSSKIGSKIG 45
+ ++L PN + + + W+ KQQQ++ K N S + IG
Sbjct 627 VEVELTPNDAKLDEDATSRDKWESKQQQEADKDCNESSVCKNIG 670
> YPR010c
Length=1203
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query 36 NSSKIGSKIGRKRRSKMRIPYGSFAAVALVGEGIIYLRRDRLV 78
NS + GRKR +R+ G AL+G G +L +DRL+
Sbjct 1053 NSLTMQPVKGRKRHGGIRV--GEMERDALIGHGTSFLLQDRLL 1093
> CE24397
Length=676
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 2/29 (6%)
Query 11 NQYQKKQQQNWQQNWQQKQQQDSSKNSSK 39
N+Y K +Q +QQN Q+ Q DS K S+
Sbjct 534 NEYMKSAKQRFQQN--QRTQSDSRKRISR 560
> At5g56320
Length=255
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 19/45 (42%), Gaps = 0/45 (0%)
Query 11 NQYQKKQQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRRSKMRIP 55
N + +NW QNWQ + D S K+ + GR S P
Sbjct 193 NTRWQSMSRNWGQNWQSNAKLDGQALSFKVTTSDGRTVISNNATP 237
> Hs4758360
Length=208
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 4 LRLQLEPNQYQKKQQQNWQQNWQQKQQQDSSKNSSKIGSKIGRKRRSKMRIP 55
L+ ++E N Y NWQ N +Q + K + + G K RK S +P
Sbjct 152 LKERIEENGYNTYASFNWQHNGRQMYVALNGKGAPRRGQKTRRKNTSAHFLP 203
Lambda K H
0.322 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40