bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3965_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
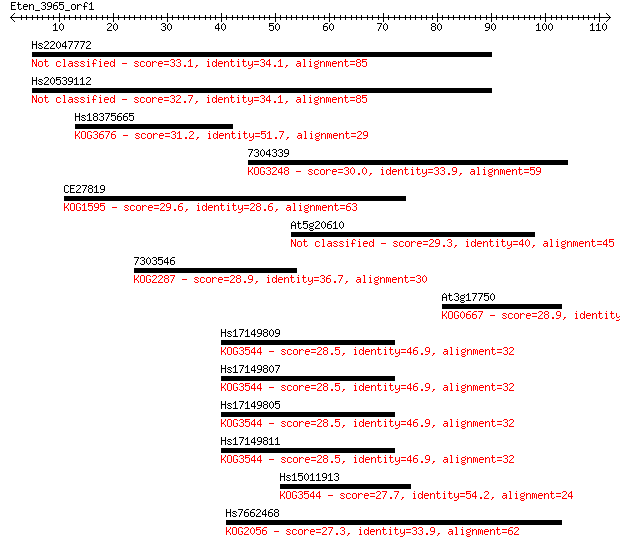
Hs22047772 33.1 0.13
Hs20539112 32.7 0.15
Hs18375665 31.2 0.48
7304339 30.0 1.0
CE27819 29.6 1.6
At5g20610 29.3 1.6
7303546 28.9 2.3
At3g17750 28.9 2.6
Hs17149809 28.5 2.8
Hs17149807 28.5 2.8
Hs17149805 28.5 2.8
Hs17149811 28.5 3.3
Hs15011913 27.7 4.8
Hs7662468 27.3 7.7
> Hs22047772
Length=777
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 35/97 (36%), Gaps = 23/97 (23%)
Query 5 GPKAYFPSPFFPHPIGGFFPTTTSLGILGGPFFFFRG------------GLGPNFFCSPG 52
GP + P P P G FP T GPF RG G +F
Sbjct 690 GP-GFIPPPLAPVR-GPLFPVDTR-----GPFMR-RGPPFPPPPPGTMFGASRGYFPPRD 741
Query 53 FPGPPGHHGGPLFNGRLFPPIKISFYFYPPLGLGSRP 89
FPGPP P ++PP + YF+P G P
Sbjct 742 FPGPPH---APFAMRNIYPPRGLPPYFHPRPGFYPNP 775
> Hs20539112
Length=777
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 35/97 (36%), Gaps = 23/97 (23%)
Query 5 GPKAYFPSPFFPHPIGGFFPTTTSLGILGGPFFFFRG------------GLGPNFFCSPG 52
GP + P P P G FP T GPF RG G +F
Sbjct 690 GP-GFIPPPLAPVR-GPLFPVDTR-----GPFMR-RGPPFPPPPPGTMFGASRGYFPPRD 741
Query 53 FPGPPGHHGGPLFNGRLFPPIKISFYFYPPLGLGSRP 89
FPGPP P ++PP + YF+P G P
Sbjct 742 FPGPPH---APFAMRNIYPPRGLPPYFHPRPGFYPNP 775
> Hs18375665
Length=839
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 1/30 (3%)
Query 13 PFFPHPIGGFFPTTTS-LGILGGPFFFFRG 41
PF IG +F T L +LGG +FFFRG
Sbjct 463 PFKMEKIGDYFRVTGEILSVLGGVYFFFRG 492
> 7304339
Length=754
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 28/62 (45%), Gaps = 16/62 (25%)
Query 45 PNFFCSPGFP---GPPGHHGGPLFNGRLFPPIKISFYFYPPLGLGSRPLIRTFGGGGWTF 101
P FFC P PP H G PP ++ P +GL +RP + F GG + +
Sbjct 112 PPFFCHNADPLSTPPPAHCG--------IPPYQLD----PKMGL-TRPALYPFAGGQYPY 158
Query 102 PL 103
P+
Sbjct 159 PM 160
> CE27819
Length=704
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 26/63 (41%), Gaps = 2/63 (3%)
Query 11 PSPFFPHPIGGFFPTTTSLGILGGPFFFFRGGLGPNFFCSPGFPGPPGHHGGPLFNGRLF 70
P+ F PI GF P+ +S +L + G S +P PG+ P+
Sbjct 326 PTERFESPINGFSPSYSS--VLKNRIPIAKQGYATTDSISSSYPKAPGYERSPIVGSFEM 383
Query 71 PPI 73
PPI
Sbjct 384 PPI 386
> At5g20610
Length=1164
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 2/45 (4%)
Query 53 FPGPPGHHGGPLFNGRLFPPIKISFYFYPPLGLGSRPLIRTFGGG 97
F P H+G F+ FP +K F PPLG G P+++T GG
Sbjct 676 FQNSPPHNGRDAFHPADFP-VKEPFDL-PPLGDGLGPVVQTKNGG 718
> 7303546
Length=1256
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 15/30 (50%), Gaps = 0/30 (0%)
Query 24 PTTTSLGILGGPFFFFRGGLGPNFFCSPGF 53
P T L P + FRGG+ P + C G+
Sbjct 746 PATNKLNKWYMPSYMFRGGVYPRYLCGSGY 775
> At3g17750
Length=1138
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 81 PPLGLGSRPLIRTFGGGGWTFP 102
PP G G+ L++ F GG++FP
Sbjct 642 PPPGKGASMLLKNFADGGFSFP 663
> Hs17149809
Length=2975
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query 40 RGGLGPNFFCSPGFPGPPGHHGGPLFNGRLFP 71
RG G F PG+PGP G+ G P NG P
Sbjct 2106 RGKKGERGF--PGYPGPKGNPGEPGLNGTTGP 2135
> Hs17149807
Length=2976
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query 40 RGGLGPNFFCSPGFPGPPGHHGGPLFNGRLFP 71
RG G F PG+PGP G+ G P NG P
Sbjct 2107 RGKKGERGF--PGYPGPKGNPGEPGLNGTTGP 2136
> Hs17149805
Length=3009
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query 40 RGGLGPNFFCSPGFPGPPGHHGGPLFNGRLFP 71
RG G F PG+PGP G+ G P NG P
Sbjct 2140 RGKKGERGF--PGYPGPKGNPGEPGLNGTTGP 2169
> Hs17149811
Length=2970
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query 40 RGGLGPNFFCSPGFPGPPGHHGGPLFNGRLFP 71
RG G F PG+PGP G+ G P NG P
Sbjct 2101 RGKKGERGF--PGYPGPKGNPGEPGLNGTTGP 2130
> Hs15011913
Length=1028
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 1/25 (4%)
Query 51 PGFPGPPGHHGGPLFNG-RLFPPIK 74
PGFPG PG+ G P NG + +P +K
Sbjct 522 PGFPGYPGNRGAPGINGTKGYPGLK 546
> Hs7662468
Length=641
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 25/63 (39%), Gaps = 7/63 (11%)
Query 41 GGLGPNFFCSPGFPGPPGHH-GGPLFNGRLFPPIKISFYFYPPLGLGSRPLIRTFGGGGW 99
GG ++ SP P H G PL P+ F P LGL SRP G W
Sbjct 190 GGSPASYHGSPEASLCPQHRTGAPLGQSEELQPLSQRHPFLPHLGLASRP------NGDW 243
Query 100 TFP 102
+ P
Sbjct 244 SQP 246
Lambda K H
0.329 0.161 0.591
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40