bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3629_orf7
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
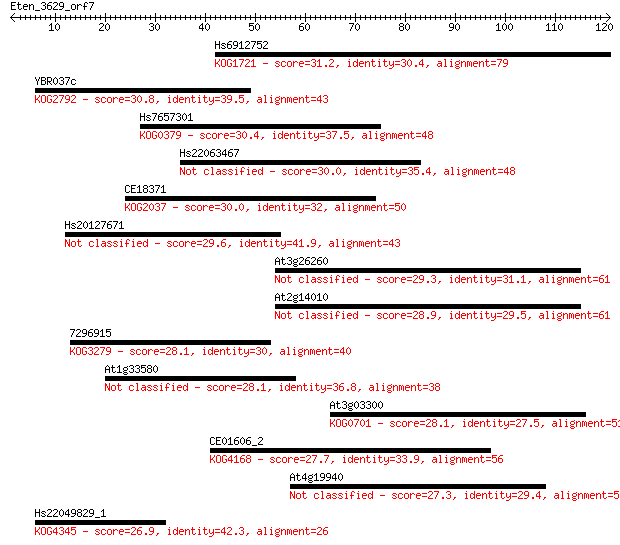
Hs6912752 31.2 0.48
YBR037c 30.8 0.63
Hs7657301 30.4 0.80
Hs22063467 30.0 1.1
CE18371 30.0 1.2
Hs20127671 29.6 1.3
At3g26260 29.3 1.8
At2g14010 28.9 2.5
7296915 28.1 3.7
At1g33580 28.1 3.7
At3g03300 28.1 4.6
CE01606_2 27.7 5.4
At4g19940 27.3 6.2
Hs22049829_1 26.9 8.0
> Hs6912752
Length=895
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 37/82 (45%), Gaps = 8/82 (9%)
Query 42 DSCKRMCTYAWETEKHRRVNTSRQ-VPC-SCAGACDRTDKH-AYTRTCWALALKGSTSRH 98
D C + E+H+R ++ + C +C RTD+ + RTC + +KG+TS
Sbjct 320 DQCSMKFIQKYHMERHKRTHSGEKPYKCDTCQQYFSRTDRLLKHRRTCGEVIVKGATSAE 379
Query 99 LSSCTHAKHQHLYGTVTAVASQ 120
S H +L AV SQ
Sbjct 380 PGSSNHTNMGNL-----AVLSQ 396
> YBR037c
Length=295
Score = 30.8 bits (68), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 21/44 (47%), Gaps = 5/44 (11%)
Query 6 LICTCDDVRDAPVPLHE-CSNTHMCIWHVYAGVSGLHDSCKRMC 48
L TCD RD+P L E S+ H I G++G D K C
Sbjct 177 LFITCDPARDSPAVLKEYLSDFHPSIL----GLTGTFDEVKNAC 216
> Hs7657301
Length=406
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 27 HMCIWHVYAG--VSGLHD---SCKRMCTYAWETEKHRRVNTSRQVPCSCAGAC 74
HM +W Y V GL+D + + Y ET + +++NT VP S +G+C
Sbjct 43 HMFVWGGYKSNQVRGLYDFYLPREELWIYNMETGRWKKINTEGDVPPSMSGSC 95
> Hs22063467
Length=428
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 3/51 (5%)
Query 35 AGVSGLHDSCKRMCTYAWETEKH---RRVNTSRQVPCSCAGACDRTDKHAY 82
+GV L D+ T + ET+ H RV + R +P +CA T H Y
Sbjct 274 SGVGSLRDTPDACATISSETKPHCYGTRVGSLRDIPDACATISSETKPHCY 324
> CE18371
Length=359
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query 24 SNTHMCIWHVYAGVSGLH--DSCKRMCTYAWETEKHRRVNTSRQVPCSCAGA 73
S + H+ G++ L+ D C R+ E EK++R+N +++V CA A
Sbjct 240 SEKKLLEAHIKHGITALNIFDKCPRIGAKEAELEKYKRLNEAKRV-TGCASA 290
> Hs20127671
Length=941
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 10/47 (21%)
Query 12 DVRDAPVPLHECSNTHMCIW----HVYAGVSGLHDSCKRMCTYAWET 54
D+ P+ L C NTHM IW + Y G+ L C AWET
Sbjct 638 DISIRPL-LEHCENTHMTIWLGIVYAYKGLLMLFG-----CFLAWET 678
> At3g26260
Length=989
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 1/61 (1%)
Query 54 TEKHRRVNTSRQVPCSCAGACDRTDKHAYTRTCWALALKGSTSRHLSSCTHAKHQHLYGT 113
+ K R+ +P S D+ DK TR A+A + L TH KH H+Y
Sbjct 373 SSKTRKAKGQFSIPDSIKNYIDKKDKRMETRMLKAIADSEARLSTLLRATHGKH-HVYEE 431
Query 114 V 114
V
Sbjct 432 V 432
> At2g14010
Length=833
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 1/61 (1%)
Query 54 TEKHRRVNTSRQVPCSCAGACDRTDKHAYTRTCWALALKGSTSRHLSSCTHAKHQHLYGT 113
+ K R+ +P S D+ DK +R A+A + L TH KH H+Y
Sbjct 339 SSKTRKAEGQFNIPDSIKNYIDKKDKRMESRMLKAIADSEARLSSLLRATHGKH-HMYEE 397
Query 114 V 114
V
Sbjct 398 V 398
> 7296915
Length=302
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 20/42 (47%), Gaps = 2/42 (4%)
Query 13 VRDAPVPLHECSNTHMCIWHVYAGVSGLH--DSCKRMCTYAW 52
V++ P+PL + +H IW A + G + KR + W
Sbjct 69 VQNIPLPLIDTPESHRGIWGGEAVIKGFQKREQTKRRVPHFW 110
> At1g33580
Length=428
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 19/38 (50%), Gaps = 4/38 (10%)
Query 20 LHECSNTHMCIWHVYAGVSGLHDSCKRMCTYAWETEKH 57
+H C N CI +Y L D+C R + WE +KH
Sbjct 219 IHACPND--CI--LYRKEYELRDTCPRCSAFRWERDKH 252
> At3g03300
Length=2042
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query 65 QVPCSCAGACDRTDKHAYTRTCWAL-ALKGSTSRHLSSCTHAKHQHLYGTVT 115
+ PC G + C+AL A+K + +H+ +H H+H+ TV+
Sbjct 1860 KYPCLSPGLLTDMRSASVNNECYALVAVKANLHKHILYASHHLHKHISRTVS 1911
> CE01606_2
Length=128
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 7/61 (11%)
Query 41 HDSCKRMCTYAWETEKHRRVNTSRQVPCSCAGACDRTDKHAYTR-----TCWALALKGST 95
H+ CK++ T +ET+K+ +NTS V + A C+ K A + T + L+ ST
Sbjct 33 HERCKQLSTVIYETKKY--LNTSPAVTQNEAVICELIPKLAPFKLTGAETLQVINLRPST 90
Query 96 S 96
+
Sbjct 91 T 91
> At4g19940
Length=411
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 23/53 (43%), Gaps = 2/53 (3%)
Query 57 HRRVNTSRQVPCSCAGACDRTDKHAYTRTCWALALKGSTSRHLS--SCTHAKH 107
H V +V C + A D +++ + W L L+G SR SC + H
Sbjct 185 HDPVYDQYKVVCIVSRASDEVEEYTFLSEHWVLLLEGEGSRRWRKISCKYPPH 237
> Hs22049829_1
Length=490
Score = 26.9 bits (58), Expect = 8.0, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 6 LICTCDDVRDAPVPLHECSNTHMCIW 31
L+C+ D V+D VP + SN C W
Sbjct 379 LVCSYDSVKDVLVPDYGMSNLTACNW 404
Lambda K H
0.324 0.129 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40