bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2433_orf2
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
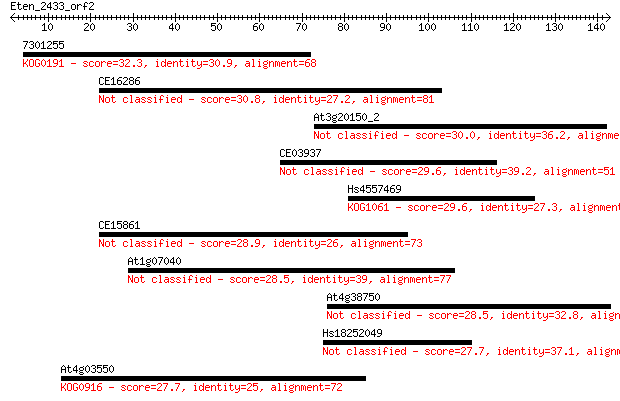
7301255 32.3 0.31
CE16286 30.8 0.98
At3g20150_2 30.0 1.3
CE03937 29.6 1.7
Hs4557469 29.6 2.0
CE15861 28.9 3.5
At1g07040 28.5 4.3
At4g38750 28.5 4.4
Hs18252049 27.7 7.0
At4g03550 27.7 7.3
> 7301255
Length=249
Score = 32.3 bits (72), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 6/68 (8%)
Query 4 VLRISMLSSGISVMRRTGIIACPLFVSATLRRVYRTQRLSRQTKSPEALRQRFVYAYLAH 63
+ R++ L SGIS R G++ P F+ ++Y + S + SPEA Q +A +
Sbjct 181 IARMNSLESGISTATRLGVLEAPAFIFLRQGKMY---KYSAKQYSPEAFVQ---FAEKGY 234
Query 64 RTKTIQPV 71
QPV
Sbjct 235 TQSHPQPV 242
> CE16286
Length=332
Score = 30.8 bits (68), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 13/83 (15%)
Query 22 IIACPLFVSATLRRVYRTQRLSRQTKSPEALRQRFVYAYLAHRTKTIQPVLSSRFARSLR 81
++ P+FV +++ V +T+ + Q F++ L H T I P ++ F R
Sbjct 255 LVYVPIFVCSSISLVTKTEYI---------FPQYFIFV-LPHLTTVIDPAVTMYFVTPYR 304
Query 82 KSVITSWKCLSNGA--TLPHTTF 102
K +I W L N ++ H+TF
Sbjct 305 KKLII-WLRLKNNKMHSIAHSTF 326
> At3g20150_2
Length=672
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 34/75 (45%), Gaps = 18/75 (24%)
Query 73 SSRFARSLRKSVITSWKCLSN----GATLPHTTF--KCHLDSNTWGSVVYDGEIFSIAVG 126
S +F SLRKS+ S CL N ++ T F H+ S+ GS ++ G S+
Sbjct 188 SPKFRDSLRKSIALSSSCLRNQNSLAKSIKSTCFAESQHIRSSLRGSKIFTGSTESL--- 244
Query 127 CVADAYFHFAASLRR 141
AASLRR
Sbjct 245 ---------AASLRR 250
> CE03937
Length=463
Score = 29.6 bits (65), Expect = 1.7, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 65 TKTIQPVLSSRFARSLRKSVITSWKCLSNGATLPHTTFKCHLDSNTWGSVV 115
+KTI L S F+ + VI KC+S +LP +T C LD N +G V
Sbjct 90 SKTIM-YLCSPFSLKRQNLVIEHQKCISGVLSLPAST-GCQLDDNEYGKQV 138
> Hs4557469
Length=937
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 81 RKSVITSWKCLSNGATLPHTTFKCHLDSNTWGSVVYDGEIFSIA 124
R+ + +WK + N L +CHL+++T S + + +++IA
Sbjct 834 RQVFLATWKDIPNENELQFQIKECHLNADTVSSKLQNNNVYTIA 877
> CE15861
Length=317
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 11/73 (15%)
Query 22 IIACPLFVSATLRRVYRTQRLSRQTKSPEALRQRFVYAYLAHRTKTIQPVLSSRFARSLR 81
++ P+F+ +T+ + TK+ Q F++ L H T I P+L+ F R
Sbjct 240 LVYVPIFICSTISLI---------TKTEYTFAQFFIFV-LPHLTTVIDPLLTMYFVTPYR 289
Query 82 KSVITSWKCLSNG 94
K ++ W L N
Sbjct 290 KRLMV-WLRLKND 301
> At1g07040
Length=371
Score = 28.5 bits (62), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 40/88 (45%), Gaps = 12/88 (13%)
Query 29 VSATLRRVYRTQ----RLSRQT-----KSPEALRQRFVYAYL--AHRTKTIQPVLSSRFA 77
V+ L RV+ ++ ++S QT K EA YA L A T T + LSS F
Sbjct 170 VNGPLPRVFISELLVDQMSSQTQDVIRKYTEASPNGKKYAGLSSALGTLTWEKPLSSEFE 229
Query 78 RSLRKSVITSWKCLSNGATLPHTTFKCH 105
+ R+S +W L NG L H T H
Sbjct 230 QLARESEYAAW-TLVNGYALNHVTISVH 256
> At4g38750
Length=1073
Score = 28.5 bits (62), Expect = 4.4, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 6/72 (8%)
Query 76 FARSLRKSVITSWKCLSNGATLPHTTFKCHLDS----NTWGSV-VYDGEIFSIAVGCVAD 130
FA SL+K + + K SNG + P T+ D + W + V DG I S++ +
Sbjct 275 FAESLKKPCVETKKTASNGVSPPKLTWTADSDPKDIFSKWCDISVLDGIIQSVS-SLDGE 333
Query 131 AYFHFAASLRRL 142
+ +F A R+L
Sbjct 334 SEINFQAKARKL 345
> Hs18252049
Length=524
Score = 27.7 bits (60), Expect = 7.0, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query 75 RFARSLRKSVITSWKCLSNGATLPHTTFKCHLDSN 109
RFA + ++++T+ S+ ATLP TF+C ++N
Sbjct 315 RFAMGMAQALLTALMISSSSATLP-VTFRCAEENN 348
> At4g03550
Length=1780
Score = 27.7 bits (60), Expect = 7.3, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 13 GISVMRRTGIIACPLFVSATLRRVYRTQRLSRQTKSPEALRQRFVYAYLAHRTKTIQPVL 72
G ++M+ T ++AC ++ S ++ + + + K EALR +V A R +T +
Sbjct 1065 GTALMKFTYVVACQIYGSQKAKKEPQAEEILYLMKQNEALRIAYVDEVPAGRGETDYYSV 1124
Query 73 SSRFARSLRKSV 84
++ L K V
Sbjct 1125 LVKYDHQLEKEV 1136
Lambda K H
0.327 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40