bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1973_orf2
Length=119
Score E
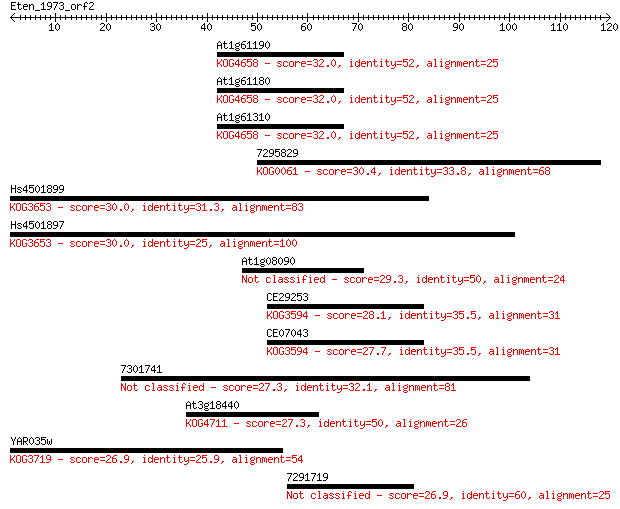
Sequences producing significant alignments: (Bits) Value
At1g61190 32.0 0.26
At1g61180 32.0 0.27
At1g61310 32.0 0.29
7295829 30.4 0.83
Hs4501899 30.0 0.99
Hs4501897 30.0 1.1
At1g08090 29.3 1.7
CE29253 28.1 3.6
CE07043 27.7 4.9
7301741 27.3 6.3
At3g18440 27.3 7.2
YAR035w 26.9 9.0
7291719 26.9 9.1
> At1g61190
Length=967
Score = 32.0 bits (71), Expect = 0.26, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 42 SRQNLNERSPAPPRAESAARPPQPT 66
S N +E S PPR+E RP QPT
Sbjct 130 SEGNFDEVSQPPPRSEVEERPTQPT 154
> At1g61180
Length=889
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 42 SRQNLNERSPAPPRAESAARPPQPT 66
S N +E S PPR+E RP QPT
Sbjct 129 SEGNFDEVSQPPPRSEVEERPTQPT 153
> At1g61310
Length=925
Score = 32.0 bits (71), Expect = 0.29, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 42 SRQNLNERSPAPPRAESAARPPQPT 66
S N +E S PPR+E RP QPT
Sbjct 131 SEGNFDEVSQPPPRSEVEERPTQPT 155
> 7295829
Length=812
Score = 30.4 bits (67), Expect = 0.83, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 4/69 (5%)
Query 50 SPAPPRAESAARPPQPTPNNLANIFPANLLQLEGQI-QRVEWLFQSPRGAVPMDPSSFVA 108
+P P + S+ RPP T N A A L + E + E ++ +P GA P+ S A
Sbjct 50 TPVPSKKLSSKRPPHLTLNIKAKSHDAPLAETENTVLSTSESVYGTPLGATPV---SSPA 106
Query 109 KHFSRSFAS 117
RS AS
Sbjct 107 NQSQRSQAS 115
> Hs4501899
Length=512
Score = 30.0 bits (66), Expect = 0.99, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 35/91 (38%), Gaps = 12/91 (13%)
Query 1 SWRNVRENAEAIAAICISEEFGAAAPTDREDSAAFR-----FCCCCSRQNLNERSPAPPR 55
SW N E + C ++F DR++ A + CCC NER P
Sbjct 62 SWANSSGTIELVKKGCWLDDFNC---YDRQECVATEENPQVYFCCCEGNFCNERFTHLPE 118
Query 56 A---ESAARPPQPTPNNLANIFPANLLQLEG 83
A E PP PT L + +LL + G
Sbjct 119 AGGPEVTYEPP-PTAPTLLTVLAYSLLPIGG 148
> Hs4501897
Length=513
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 25/108 (23%), Positives = 44/108 (40%), Gaps = 9/108 (8%)
Query 1 SWRNVRENAEAIAAICISEEFGAAAPTD---REDSAAFRFCCCCSRQNLNERSPAPPRAE 57
+W+N+ + E + C ++ TD ++DS F CCC NE+ P E
Sbjct 63 TWKNISGSIEIVKQGCWLDDINCYDRTDCVEKKDSPEVYF-CCCEGNMCNEKFSYFPEME 121
Query 58 -----SAARPPQPTPNNLANIFPANLLQLEGQIQRVEWLFQSPRGAVP 100
S P+P N+ L+ + G + W+++ + A P
Sbjct 122 VTQPTSNPVTPKPPYYNILLYSLVPLMLIAGIVICAFWVYRHHKMAYP 169
> At1g08090
Length=530
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 47 NERSPAPPRAESAARPPQPTPNNL 70
N +S R SAA PP+ TPNN+
Sbjct 507 NAKSEGGRRVRSAATPPENTPNNV 530
> CE29253
Length=1306
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 52 APPRAESAARPPQPTPNNLANIFPANLLQLE 82
APP+ PP PTP LA +F + + ++
Sbjct 537 APPQPVPTQSPPLPTPKRLAPVFKPSQITVQ 567
> CE07043
Length=1544
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 52 APPRAESAARPPQPTPNNLANIFPANLLQLE 82
APP+ PP PTP LA +F + + ++
Sbjct 681 APPQPVPTQSPPLPTPKRLAPVFKPSQITVQ 711
> 7301741
Length=2977
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 35/85 (41%), Gaps = 4/85 (4%)
Query 23 AAAPTDREDSAAFRFCCCCSRQNLNE--RSPAPPRAESAARPPQPTPNNLANIFPANLL- 79
+ APT DS F+ + L ++ P A S+A+PP PTP A +L
Sbjct 1139 SVAPTITSDSTGFKLSSDGRQMQLVAIPQATPPSPAASSAQPPVPTPTPALTAGGATILP 1198
Query 80 -QLEGQIQRVEWLFQSPRGAVPMDP 103
QL G V SP A + P
Sbjct 1199 PQLTGMPANVSVSVGSPASAAALVP 1223
> At3g18440
Length=598
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 1/27 (3%)
Query 36 RFCCCCSRQNLNER-SPAPPRAESAAR 61
R CCCCS NL+E+ S A+ AR
Sbjct 49 RLCCCCSCGNLSEKISGVYDDAKDVAR 75
> YAR035w
Length=687
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 24/54 (44%), Gaps = 3/54 (5%)
Query 1 SWRNVRENAEAIAAICISEEFGAAAPTDREDSAAFRFCCCCSRQNLNERSPAPP 54
+WRN++ A+ +C+ + A D++D C S NL+ PP
Sbjct 264 NWRNLKLIDSALFVVCLDD---VAFAADQQDELTRSMLCGTSTINLDPHQHQPP 314
> 7291719
Length=333
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 7/32 (21%)
Query 56 AESAARPPQPT-------PNNLANIFPANLLQ 80
AES +PP+PT PNN AN F AN+ +
Sbjct 135 AESLHKPPKPTADSIKMSPNNSANEFCANIFE 166
Lambda K H
0.319 0.130 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40