bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1775_orf5
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
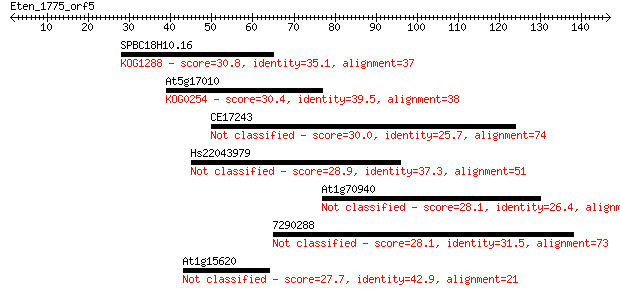
SPBC18H10.16 30.8 1.00
At5g17010 30.4 1.3
CE17243 30.0 1.4
Hs22043979 28.9 4.0
At1g70940 28.1 6.3
7290288 28.1 6.3
At1g15620 27.7 7.3
> SPBC18H10.16
Length=1050
Score = 30.8 bits (68), Expect = 1.00, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 28 LQQVSPESFIFLLLFFIKRLAICCCCCCCSFATFCFA 64
+ Q PE + ++ L+ LA+CC CC A F A
Sbjct 194 ISQFLPEGYWWVFLYTTCVLAMCCILCCLGSAIFAKA 230
> At5g17010
Length=440
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 39 LLLFFIKRLAICCCCCCCSFATFCFAFPRI-PQRIAGRG 76
LLL + + + CCC C+ A C P I P ++ GRG
Sbjct 337 LLLGGVGGMRLTSCCCSCT-AALCGLLPEIFPLKLRGRG 374
> CE17243
Length=553
Score = 30.0 bits (66), Expect = 1.4, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 31/74 (41%), Gaps = 9/74 (12%)
Query 50 CCCCCCCSFATFCFAFPRIPQRIAGRGGGGCSCSHSAQANLNWNLNSNLNLNLNLNFSEL 109
C C C+ + R A R GGC+C+H+ AN + +L +
Sbjct 57 CTACSQCTENGYSVCL-RPKDSRASRTAGGCTCAHTLAANCHTDLAQTQH--------PA 107
Query 110 NSNSKSYLNCKANS 123
N + K+Y C A++
Sbjct 108 NLSMKTYSACYADA 121
> Hs22043979
Length=221
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 23/53 (43%), Gaps = 5/53 (9%)
Query 45 KRLAICCCCCCCSFATFCFAFPRIPQRIAG--RGGGGCSCSHSAQANLNWNLN 95
K L C CS CF P P+R+ G GGG H A A+ WN +
Sbjct 9 KHLVETECELSCS--ELCFRLPPQPRRLLGLQVSGGGLKLQHPA-ADTFWNFS 58
> At1g70940
Length=640
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 77 GGGCSCSHSAQANLNWNLNSNLNLNLNLNFSELNSNSKSYLNCKANSNSQDKR 129
GGG + + S N ++N N ++N+N ++ L SNS D +
Sbjct 302 GGGAGSYPAPNPEFSSTTTSTANKSVNKNPKDVNTNQQTTLPTGGKSNSHDAK 354
> 7290288
Length=715
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 37/74 (50%), Gaps = 3/74 (4%)
Query 65 FPRIPQRIAGRGGGGCSC-SHSAQANLNWNLNSNLNLNLNLNFSELNSNSKSYLNCKANS 123
+ ++PQR GG C S+S NL NLN+N N +N L + ++ + K+ S
Sbjct 347 YLQLPQRTDTAVSGGALCRSNSVGHNLKPNLNNNSNGAVNGQGPTLVTPPQAVV--KSTS 404
Query 124 NSQDKRGTNSESKL 137
N R +S+ +L
Sbjct 405 NDDIARKCHSQLQL 418
> At1g15620
Length=343
Score = 27.7 bits (60), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 9/21 (42%), Positives = 12/21 (57%), Gaps = 0/21 (0%)
Query 43 FIKRLAICCCCCCCSFATFCF 63
++ + IC CCC F T CF
Sbjct 318 YVLPMVICAATCCCVFMTLCF 338
Lambda K H
0.325 0.137 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40