bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0763_orf1
Length=133
Score E
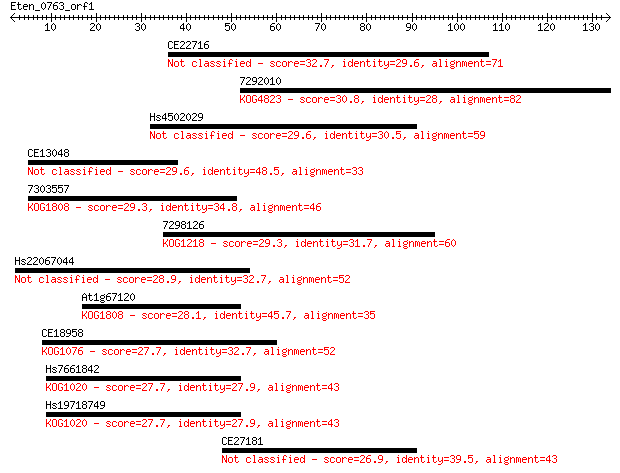
Sequences producing significant alignments: (Bits) Value
CE22716 32.7 0.18
7292010 30.8 0.67
Hs4502029 29.6 1.8
CE13048 29.6 1.8
7303557 29.3 2.1
7298126 29.3 2.4
Hs22067044 28.9 2.8
At1g67120 28.1 4.6
CE18958 27.7 5.5
Hs7661842 27.7 5.5
Hs19718749 27.7 5.6
CE27181 26.9 9.8
> CE22716
Length=846
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Query 36 EEGGGQEEKQEQEGENNNW---AIAGGVVGGLILLAAVAAGAVFYTKKPSSQAQDSHVVI 92
EE G+ EK ++E W AI GG + ++ + +V AG Y KK +++ + S++ +
Sbjct 398 EESDGKAEKDKKEDTTWTWHTYAITGGAIIAILFILSVCAGLKCY-KKFNNKKKASNIHL 456
Query 93 VEEHGTGGGAGRTP 106
+ E+ +G P
Sbjct 457 LNENPAFSHSGSIP 470
> 7292010
Length=735
Score = 30.8 bits (68), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 4/82 (4%)
Query 52 NNWAIAGGVVGGLILLAAVAAGAVFYTKKPSSQAQDSHVVIVEEHGTGGGAGRTPSEANV 111
NN+A G +V G VA+ + + + S+ + VV GGG+ + PS +N
Sbjct 99 NNFAYVGPIVMGFGGFIVVASCVMTFEARDSA----AKVVPARLRAGGGGSSKNPSRSNQ 154
Query 112 SLEESFWRGDGEGLIDANYDQQ 133
S RG G + +DQ
Sbjct 155 GSSASQRRGMGAQYQSSRWDQH 176
> Hs4502029
Length=583
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 32 SLPPEEGGGQEEKQEQEGENNNWAIAGGVVGGLILLAAVAAGAVFYTKKPSSQAQDSHV 90
S+P + + + +E N+ + G+V GL LLAA+ AG V++ S+ HV
Sbjct 504 SIPEHDEADEISDENREKVNDQAKLIVGIVVGL-LLAALVAGVVYWLYMKKSKTASKHV 561
> CE13048
Length=585
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 4/37 (10%)
Query 5 EPGEPGSGEGGGEEGET-NPPKEPQEEPS---LPPEE 37
+P EPG+ E EGE NP +E +E P+ LP E+
Sbjct 107 KPPEPGNRENANVEGEDGNPVEEDEEMPAHEDLPSED 143
> 7303557
Length=4865
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 5 EPGEPGSGEGGGEEGETNPPKEPQEEPSLPPEEGGGQEEKQEQEGE 50
E G+P S EE E +P EE EE ++EK E E +
Sbjct 4642 EEGDPQSDGSDSEEDEAGTEAKPAEEDHGEGEEATPEDEKDEAETQ 4687
> 7298126
Length=543
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 35 PEEGGGQEEK---QEQEGENNNWAIAGGVVGGLIL-LAAVAAGAVFYTKKPSSQAQDSHV 90
PE+G E + ++ G +N W I G G+I+ +AA G +Y + S + ++
Sbjct 476 PEQGARSEAEVRIPDRTGSSNKWMIIVGSCAGMIIGVAATIIGIKYYRRSTSRRNFEAEE 535
Query 91 VIVE 94
IVE
Sbjct 536 AIVE 539
> Hs22067044
Length=551
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 22/53 (41%), Gaps = 1/53 (1%)
Query 2 PSPEPGEPGSGEGGGEEGETNPPKE-PQEEPSLPPEEGGGQEEKQEQEGENNN 53
P P PG + +G+ PPKE P +P +E EGEN N
Sbjct 218 PGESPRPPGGPQAHPRKGQAPPPKETPSPVKGMPNPAEKNSGPAKECEGENAN 270
> At1g67120
Length=5138
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query 17 EEGETNPPKEPQEEPSLPPEEGGG-QEEKQEQEGEN 51
E+ + N +EP E PEEG QEE QE EN
Sbjct 4496 EKEDANQQEEPCSEDQKHPEEGENDQEETQEPSEEN 4531
> CE18958
Length=898
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 25/55 (45%), Gaps = 3/55 (5%)
Query 8 EPGSGEGGGEEGETNPPKEPQEEPSLPPEEGGGQEEK---QEQEGENNNWAIAGG 59
EP +G GG + + P + + GG Q E+ Q Q+G+ NW GG
Sbjct 832 EPRTGRGGYQGPGSWFPGRNERQGDKQKGSGGYQGERRGGQGQDGKRGNWGSQGG 886
> Hs7661842
Length=2158
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 PGSGEGGGEEGETNPPKEPQEEPSLPPEEGGGQEEKQEQEGEN 51
P + + GE P ++ P P ++G G+ E +Q+GE+
Sbjct 161 PETPKQKGESRPETPKQKSDGHPETPKQKGDGRPETPKQKGES 203
> Hs19718749
Length=2265
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 PGSGEGGGEEGETNPPKEPQEEPSLPPEEGGGQEEKQEQEGEN 51
P + + GE P ++ P P ++G G+ E +Q+GE+
Sbjct 161 PETPKQKGESRPETPKQKSDGHPETPKQKGDGRPETPKQKGES 203
> CE27181
Length=703
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 20/43 (46%), Gaps = 3/43 (6%)
Query 48 EGENNNWAIAGGVVGGLILLAAVAAGAVFYTKKPSSQAQDSHV 90
E N W I GGVV ILL + AVF + S+ S V
Sbjct 199 EASNMVWYIIGGVV---ILLLVIVGIAVFLIMRKKSKPSSSEV 238
Lambda K H
0.301 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40