bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_7096_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
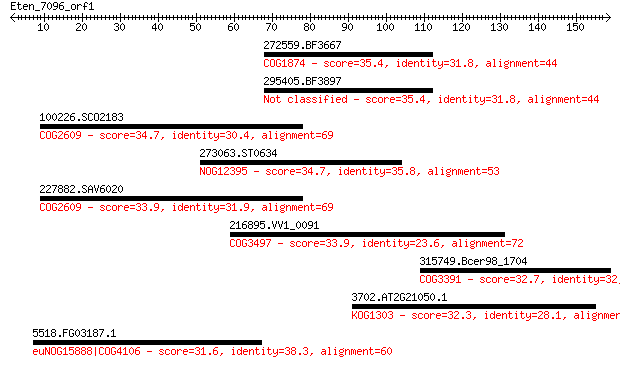
272559.BF3667 35.4 0.59
295405.BF3897 35.4 0.59
100226.SCO2183 34.7 1.00
273063.ST0634 34.7 1.0
227882.SAV6020 33.9 1.6
216895.VV1_0091 33.9 1.6
315749.Bcer98_1704 32.7 3.7
3702.AT2G21050.1 32.3 4.4
5518.FG03187.1 31.6 7.1
> 272559.BF3667
Length=769
Score = 35.4 bits (80), Expect = 0.59, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 28/44 (63%), Gaps = 9/44 (20%)
Query 68 RYSSDGQTKRGNCTGDEMFGQPKLGSWNIIEQSSYYLTVSSRSF 111
RY+ +T+ GNCTGD++F +++S+Y++TV+ ++
Sbjct 690 RYADSEETRSGNCTGDKVFD---------LQESTYWMTVAKDAY 724
> 295405.BF3897
Length=769
Score = 35.4 bits (80), Expect = 0.59, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 28/44 (63%), Gaps = 9/44 (20%)
Query 68 RYSSDGQTKRGNCTGDEMFGQPKLGSWNIIEQSSYYLTVSSRSF 111
RY+ +T+ GNCTGD++F +++S+Y++TV+ ++
Sbjct 690 RYADSEETRSGNCTGDKVFD---------LQESTYWMTVAKDAY 724
> 100226.SCO2183
Length=899
Score = 34.7 bits (78), Expect = 1.00, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 9 DESFKKWVKGIQAKKQSELLEAKDKDYQTKSAAYAQQ---MRNAVLLCKQKLQLLDEIMP 65
DE F+ G ++ E+ +A+ + YQT+ AAY ++ ++ L KL D+I+
Sbjct 307 DELFQLDTTGALVRRLREVPDAQVQTYQTRDAAYIREDFFNKDPQLAEMAKLLSDDKILD 366
Query 66 CYRYSSDGQTKR 77
C+ +S G R
Sbjct 367 CFHFSRGGHESR 378
> 273063.ST0634
Length=197
Score = 34.7 bits (78), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query 51 LLCKQKLQLLDEIMPCYRYSSDGQTKRGNCTGDEMFGQPK---LGSWNIIEQSSYY 103
L+ K+ +LL +P Y Y DG+ K+ C G +F P + +IIE S+Y
Sbjct 3 LITKRVEELLVPPLPEYSYICDGEIKQSECKGSMIFRDPDYMLITPQDIIESFSFY 58
> 227882.SAV6020
Length=900
Score = 33.9 bits (76), Expect = 1.6, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 9 DESFKKWVKGIQAKKQSELLEAKDKDYQTKSAAYAQQ---MRNAVLLCKQKLQLLDEIMP 65
DE F+ G ++ E+ +A+ + YQT+ AAY +Q + L+ KL D+I+
Sbjct 306 DELFQLDTTGALVRRLREVPDAQVQTYQTRDAAYIRQDFFGADPALVEMAKLIGDDKILE 365
Query 66 CYRYSSDGQTKR 77
C+ S G R
Sbjct 366 CFHLSRGGHESR 377
> 216895.VV1_0091
Length=386
Score = 33.9 bits (76), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Query 59 LLDEIMPCYRYSSDGQTKRGNCTGDEMFGQPKLGSWNIIEQSSYYL--TVSSRSFNQTIQ 116
+++++ + DG+T+ G+ G + + P L +I ++YL + Q I
Sbjct 312 FVEDVVESVQNGLDGETRAGHMYGAKAWADPDLNPPEVIISGNFYLDYDFTPPGLAQAIH 371
Query 117 FASSHTIDYVNALF 130
S T DY + +F
Sbjct 372 VTSHFTNDYADVIF 385
> 315749.Bcer98_1704
Length=689
Score = 32.7 bits (73), Expect = 3.7, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 30/50 (60%), Gaps = 3/50 (6%)
Query 109 RSFNQTIQFASSHTIDYVNALFALMQKVLKNLSDALSCSFVREGMIQTTY 158
R+ NQ I + ++ ++ALF Q + +NLS +SCS + + ++Q T+
Sbjct 98 RTINQLINSLPTASLTQISALF---QFLFQNLSSLISCSNLPDSLLQQTF 144
> 3702.AT2G21050.1
Length=483
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query 91 LGSWNIIEQSSYYLTVSSRSFNQTIQFASSHTIDYVNALFALMQKVLKNLSDALSCSFVR 150
LGSW S Y+ +R + + F +H I + L L+ K +N+ A +C+F+
Sbjct 83 LGSWTAYLISILYVEYRTRKEREKVNF-RNHVIQWFEVLDGLLGKHWRNVGLAFNCTFLL 141
Query 151 EGMI 154
G +
Sbjct 142 FGSV 145
> 5518.FG03187.1
Length=781
Score = 31.6 bits (70), Expect = 7.1, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 29/67 (43%), Gaps = 7/67 (10%)
Query 7 ENDESFKKWVKGIQAKKQSELL-EAKDKDY------QTKSAAYAQQMRNAVLLCKQKLQL 59
EN E +WVKG + + L E K+Y + K A AQ A C+QK
Sbjct 203 ENHEGIVEWVKGTGLRPYVDPLDENAKKEYIETYLGKLKKAYKAQNDGKACEYCRQKKTR 262
Query 60 LDEIMPC 66
DE PC
Sbjct 263 CDEDFPC 269
Lambda K H
0.315 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 25621323489
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40