bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_5425_orf5
Length=60
Score E
Sequences producing significant alignments: (Bits) Value
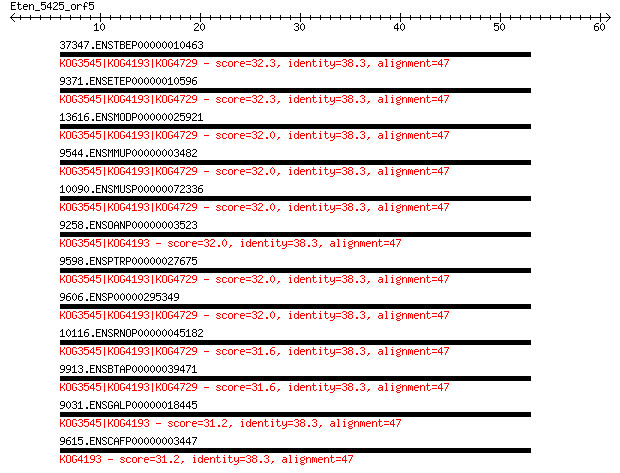
37347.ENSTBEP00000010463 32.3 4.4
9371.ENSETEP00000010596 32.3 4.4
13616.ENSMODP00000025921 32.0 5.4
9544.ENSMMUP00000003482 32.0 5.4
10090.ENSMUSP00000072336 32.0 5.5
9258.ENSOANP00000003523 32.0 5.6
9598.ENSPTRP00000027675 32.0 5.6
9606.ENSP00000295349 32.0 5.7
10116.ENSRNOP00000045182 31.6 6.8
9913.ENSBTAP00000039471 31.6 6.8
9031.ENSGALP00000018445 31.2 8.5
9615.ENSCAFP00000003447 31.2 9.2
> 37347.ENSTBEP00000010463
Length=1423
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 824 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 870
> 9371.ENSETEP00000010596
Length=1214
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 825 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 871
> 13616.ENSMODP00000025921
Length=1450
Score = 32.0 bits (71), Expect = 5.4, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 851 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 897
> 9544.ENSMMUP00000003482
Length=1429
Score = 32.0 bits (71), Expect = 5.4, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 830 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 876
> 10090.ENSMUSP00000072336
Length=1543
Score = 32.0 bits (71), Expect = 5.5, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 929 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 975
> 9258.ENSOANP00000003523
Length=1307
Score = 32.0 bits (71), Expect = 5.6, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 710 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 756
> 9598.ENSPTRP00000027675
Length=1448
Score = 32.0 bits (71), Expect = 5.6, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 849 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 895
> 9606.ENSP00000295349
Length=1448
Score = 32.0 bits (71), Expect = 5.7, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 849 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 895
> 10116.ENSRNOP00000045182
Length=1550
Score = 31.6 bits (70), Expect = 6.8, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 928 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 974
> 9913.ENSBTAP00000039471
Length=1580
Score = 31.6 bits (70), Expect = 6.8, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 929 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 975
> 9031.ENSGALP00000018445
Length=1355
Score = 31.2 bits (69), Expect = 8.5, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 756 MAHVEVKHSD-AVHDLLLDFITWVGILLSLVCLLICIFTFCFFRGLQS 802
> 9615.ENSCAFP00000003447
Length=1043
Score = 31.2 bits (69), Expect = 9.2, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 6 LCHVRLQGSQFAAVELLLSALCSVGLLLLLTTLILCTFSHC-LQGLSA 52
+ HV ++ S A +LLL + VG+LL L L++C F+ C +GL +
Sbjct 435 MAHVEVKHSD-AVHDLLLDVITWVGILLSLVCLLICIFTFCFFRGLQS 481
Lambda K H
0.327 0.140 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22504648851
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40