bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_1902_orf3
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
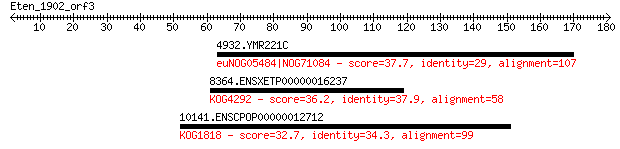
4932.YMR221C 37.7 0.18
8364.ENSXETP00000016237 36.2 0.53
10141.ENSCPOP00000012712 32.7 5.3
> 4932.YMR221C
Length=504
Score = 37.7 bits (86), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 49/116 (42%), Gaps = 18/116 (15%)
Query 63 NWIP------FLSFYEACTCAVSSCQLPSPPTQGNQVAQHRKVMRSVLVKPCGRMFGLRQ 116
NW P F + Y + +CQL P + H + + V+ GL +
Sbjct 177 NWFPTLNVSRFFTLYLIVPVFILACQLTIMPHSSYKTVNH---IAKIAVE------GLDE 227
Query 117 NGKLKRGILCLGAALKQQLRVRLGAVEREERRLGPEPMYKETKFFVF---KLERKS 169
NG+L G G +Q R L A+EREE + P +++ + KL++KS
Sbjct 228 NGRLIEGDTGSGIIPDEQERQSLIAIEREEDSIPSRPQRRKSVLETYVEDKLQKKS 283
> 8364.ENSXETP00000016237
Length=3200
Score = 36.2 bits (82), Expect = 0.53, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 26/60 (43%), Gaps = 5/60 (8%)
Query 61 DINWI--PFLSFYEACTCAVSSCQLPSPPTQGNQVAQHRKVMRSVLVKPCGRMFGLRQNG 118
D+ W+ P S ACT V C LP+PP N Q M S PC G + NG
Sbjct 213 DVGWMSPPGSS---ACTADVDECALPNPPCSQNPPVQCINTMGSFTCGPCPTDKGWQGNG 269
> 10141.ENSCPOP00000012712
Length=421
Score = 32.7 bits (73), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 50/107 (46%), Gaps = 18/107 (16%)
Query 52 HEIIHRGSSDIN---WIPFLSFYEACTCAVSSCQLPSPPTQGNQVAQHRKVMRSVLVKPC 108
HEI+ RGSS N W P + Y+ + + Q PSP +QG ++Q +V+ L +
Sbjct 107 HEILTRGSSPANASKWSPPQN-YKKRVATLEAKQKPSPQSQG--LSQQDQVIAERLAR-- 161
Query 109 GRMFGLRQNGKLKR-----GILCLGAALKQQLRVRLGAVEREERRLG 150
LRQ K K I AALK + R + +++ E RL
Sbjct 162 -----LRQENKPKSVPSQAEIEARLAALKDEPRGPITSIQEMETRLA 203
Lambda K H
0.326 0.141 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 37047630060
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40