bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_0562_orf6
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
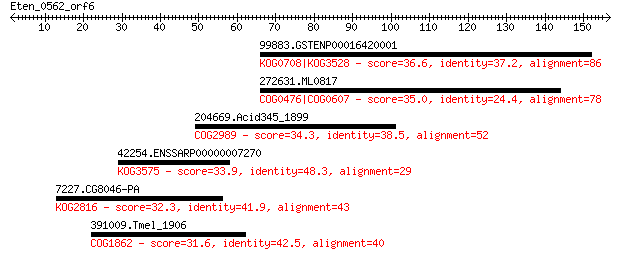
99883.GSTENP00016420001 36.6 0.26
272631.ML0817 35.0 0.66
204669.Acid345_1899 34.3 1.1
42254.ENSSARP00000007270 33.9 1.5
7227.CG8046-PA 32.3 4.4
391009.Tmel_1906 31.6 8.0
> 99883.GSTENP00016420001
Length=1830
Score = 36.6 bits (83), Expect = 0.26, Method: Composition-based stats.
Identities = 32/93 (34%), Positives = 40/93 (43%), Gaps = 10/93 (10%)
Query 66 PRCPSFCCCCCDARPLAAAAA------AAAALGSSVRAKQLLGFLGPLAARRLLLRAAR- 118
PR P CC A P A+A A L S+ Q L L P++ L R+AR
Sbjct 985 PRSPGTCCAPPPAAPTGTASALSPPSTMAPYLPVSLLQSQTLA-LAPVS--NLAPRSARF 1041
Query 119 CTYTAVRLLRAPPRGRLQQLQRQQQQQQQQQLP 151
CT + L R R +L R Q + QQLP
Sbjct 1042 CTQGPLPLSRRDSRDEESRLHRTQSPRSHQQLP 1074
> 272631.ML0817
Length=395
Score = 35.0 bits (79), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 19/78 (24%), Positives = 34/78 (43%), Gaps = 0/78 (0%)
Query 66 PRCPSFCCCCCDARPLAAAAAAAAALGSSVRAKQLLGFLGPLAARRLLLRAARCTYTAVR 125
P P C + L A+ A++ + K + G PL R ++ A +Y +R
Sbjct 197 PPPPGMVPSCAEGGVLGIVCASIASVMGTEAIKLITGIGAPLLGRLMIYNALEMSYRRIR 256
Query 126 LLRAPPRGRLQQLQRQQQ 143
+ + P R ++ +L QQ
Sbjct 257 IHKDPSRPKITELTDYQQ 274
> 204669.Acid345_1899
Length=561
Score = 34.3 bits (77), Expect = 1.1, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 11/52 (21%)
Query 49 FLLLLLFLLLLPQEAHLPRCPSFCCCCCDARPLAAAAAAAAALGSSVRAKQL 100
F+ LL+F LLLP AH + +P +A A + AL S++RA QL
Sbjct 10 FVYLLMFGLLLPHVAHAQK-----------KPASAKPAGSPALQSAIRAGQL 50
> 42254.ENSSARP00000007270
Length=549
Score = 33.9 bits (76), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 29 GRGAPRRSLPQQQQLLLLLLFLLLLLFLL 57
GRG P +LP Q Q+L+L F+ +L+FL
Sbjct 521 GRGVPVHTLPHQPQVLILAGFVFILIFLF 549
> 7227.CG8046-PA
Length=519
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 13 ELRRASGPAAGCTDTSGRGAPRRSLPQQQQLLLLLLFLLLLLF 55
E +R+SG +A T G P+ S PQ + LL F+L+LLF
Sbjct 31 EKQRSSGASAIAVGTEFPGNPQASRPQSLGMYLLEPFILILLF 73
> 391009.Tmel_1906
Length=116
Score = 31.6 bits (70), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 5/45 (11%)
Query 22 AGCTDTSGRGAPRRSLPQQ-----QQLLLLLLFLLLLLFLLLLPQ 61
AG D S G + P Q L LLL+F +++ FL++LPQ
Sbjct 7 AGVPDPSAAGNTTSAAPSAASGLFQMLFLLLIFFVMMYFLVILPQ 51
Lambda K H
0.331 0.141 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 24371502831
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40