bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8689_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
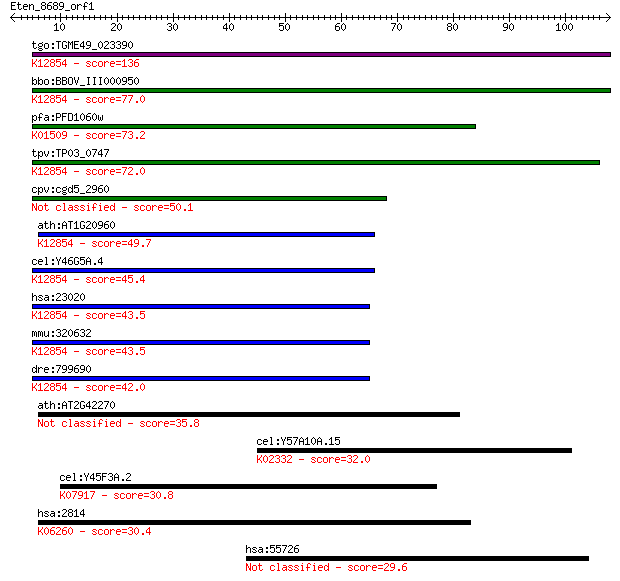
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 136 1e-32
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 77.0 1e-14
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 73.2 2e-13
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 72.0 4e-13
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 50.1 2e-06
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 49.7 2e-06
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 45.4 4e-05
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 43.5 2e-04
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 43.5 2e-04
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 42.0 5e-04
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 35.8 0.035
cel:Y57A10A.15 hypothetical protein; K02332 DNA polymerase gam... 32.0 0.44
cel:Y45F3A.2 rab-30; RAB family member (rab-30); K07917 Ras-re... 30.8 1.0
hsa:2814 GP5, CD42d; glycoprotein V (platelet); K06260 platele... 30.4 1.3
hsa:55726 C12orf11, FLJ10630, FLJ10637, NET48; chromosome 12 o... 29.6 2.3
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 136 bits (343), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 74/104 (71%), Positives = 80/104 (76%), Gaps = 8/104 (7%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPMGDKAER 64
MAE FERFKRFEYRQNSNLVLQRD TG H GEPTGEPESLAGRKLYPMGDK ER
Sbjct 1 MAEEFERFKRFEYRQNSNLVLQRDTS-TGVAPAVHLGEPTGEPESLAGRKLYPMGDKVER 59
Query 65 GLKKEEK-EKIGKRRNKLKEGGIAKRAKLDIKKGSTVLDADVTE 107
GLKKE++ K +R KL KR KLD+K+G+TVLDADVTE
Sbjct 60 GLKKEDRPVKNDSKRAKL------KRNKLDLKRGATVLDADVTE 97
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 47/108 (43%), Positives = 63/108 (58%), Gaps = 15/108 (13%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPT-GEPESLAGRKLYPMGDKAE 63
MAE +ERFKRFEYR NSNLVLQ D G +G T GEPESLA R Y MGD
Sbjct 1 MAEQYERFKRFEYRTNSNLVLQHD-------GHVSKGAVTSGEPESLANRLKYKMGD--- 50
Query 64 RGLKKEEKEKIGKRRNKLKEGGIA----KRAKLDIKKGSTVLDADVTE 107
R + + + K+R++ + +R +LD+KKG +VL D+++
Sbjct 51 RVVHSKPADLPAKKRSQASDSSSLPLKHRRTRLDLKKGDSVLTVDISD 98
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 40/86 (46%), Positives = 51/86 (59%), Gaps = 13/86 (15%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPMGDKAER 64
MAE +E+FKRFEYR NSNLVLQR+ ++ EPTGE ESL GR Y MGDK E
Sbjct 1 MAEEYEKFKRFEYRMNSNLVLQREGPISNTK------EPTGESESLVGRLKYKMGDKVEY 54
Query 65 GLKK-------EEKEKIGKRRNKLKE 83
K ++ +++ RNK K+
Sbjct 55 SNKNKKNIMLVQDNKEMAINRNKRKD 80
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 48/104 (46%), Positives = 60/104 (57%), Gaps = 12/104 (11%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPMGDKA-E 63
MAE F RF RF YR NSNLV+QR+ AP E TGEPESLAGR L+ MGDK
Sbjct 1 MAEHFVRFNRFNYRNNSNLVIQRE---GPAP---RLDESTGEPESLAGRILHKMGDKVMH 54
Query 64 RGLKKEEKEKIGKRRNKLKEGGIAKRAK--LDIKKGSTVLDADV 105
+K + K KR + + ++R K LDI +G VL+ D+
Sbjct 55 EAIKPKLKN---KRAAQTVQDEPSRRRKTFLDISRGENVLNIDI 95
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 36/63 (57%), Gaps = 7/63 (11%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPMGDKAER 64
MA+ +E KRF Y+ NSNLV+Q + + PTGE ESLAGR Y MGD A +
Sbjct 1 MADDYEMSKRFGYKANSNLVIQTSQR-------SRENVPTGEAESLAGRIKYKMGDLANK 53
Query 65 GLK 67
K
Sbjct 54 RAK 56
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 38/62 (61%), Gaps = 9/62 (14%)
Query 6 AEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYP--MGDKAE 63
AEA RFK++EYR NS+LVL D P H EPTGEPE+L G K+ P GD+
Sbjct 8 AEAHARFKQYEYRANSSLVLTTD----NRPRDTH--EPTGEPETLWG-KIDPRSFGDRVA 60
Query 64 RG 65
+G
Sbjct 61 KG 62
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 38/63 (60%), Gaps = 5/63 (7%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKL--YPMGDKA 62
MA+ R +++EYRQNSNLVL D LT G + EPTGE + +++ MGD+A
Sbjct 1 MADELARIQQYEYRQNSNLVLSVDYNLTDRRG---REEPTGEVLPITDKEMRKMKMGDRA 57
Query 63 ERG 65
+G
Sbjct 58 IKG 60
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGR-KLYPMGDKAE 63
MA+ R ++EY+ NSNLVLQ D L + EPTGE SL G+ + MGDKA+
Sbjct 1 MADVTARSLQYEYKANSNLVLQADRSLIDR---TRRDEPTGEVLSLVGKLEGTRMGDKAQ 57
Query 64 R 64
R
Sbjct 58 R 58
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGR-KLYPMGDKAE 63
MA+ R ++EY+ NSNLVLQ D L + EPTGE SL G+ + MGDKA+
Sbjct 1 MADVTARSLQYEYKANSNLVLQADRSLIDR---TRRDEPTGEVLSLVGKLEGTRMGDKAQ 57
Query 64 R 64
R
Sbjct 58 R 58
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/61 (42%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 5 MAEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGR-KLYPMGDKAE 63
MA+ R ++EY+ NSNLVLQ D L + EPTGE SL G+ + MGD+++
Sbjct 1 MADVTARSLQYEYKANSNLVLQADRSLIDR---TRRDEPTGEVLSLVGKLEGTKMGDRSQ 57
Query 64 R 64
R
Sbjct 58 R 58
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 40/77 (51%), Gaps = 9/77 (11%)
Query 6 AEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYP--MGDKAE 63
AE R K++ Y+ NS+LVL D H E +GEPESL GR + P GD+
Sbjct 9 AEEQARLKQYGYKVNSSLVLNSDE----RRRDTH--ESSGEPESLRGR-IDPKSFGDRVV 61
Query 64 RGLKKEEKEKIGKRRNK 80
RG E E++ K + K
Sbjct 62 RGRPHELDERLNKSKKK 78
> cel:Y57A10A.15 hypothetical protein; K02332 DNA polymerase gamma
1 [EC:2.7.7.7]
Length=1072
Score = 32.0 bits (71), Expect = 0.44, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 45 GEPESLAGRKLYPMGDKAERGLKKEEKEKIGKRRNKLKEGGIAKRAKLDIKKGSTV 100
G E+ AG+ L +G GLK+ E E + KL +G +AK K+D++ V
Sbjct 763 GSGETHAGKHLMRVG-----GLKQSEAESTASQLFKLTKGDVAKYMKVDVRMNCVV 813
> cel:Y45F3A.2 rab-30; RAB family member (rab-30); K07917 Ras-related
protein Rab-30
Length=216
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 37/72 (51%), Gaps = 7/72 (9%)
Query 10 ERFKRFE---YRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPM--GDKAER 64
ERF+ YR +VL D ++ P E GE ES A R++ + G+K ++
Sbjct 67 ERFRSITQSYYRSAHAIVLVYD--VSCQPSFDCLPEWLGEIESYANRRVLKILVGNKVDK 124
Query 65 GLKKEEKEKIGK 76
G ++E E+IG+
Sbjct 125 GDEREVPERIGR 136
> hsa:2814 GP5, CD42d; glycoprotein V (platelet); K06260 platelet
glycoprotein V
Length=560
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 33/77 (42%), Gaps = 7/77 (9%)
Query 6 AEAFERFKRFEYRQNSNLVLQRDPYLTGAPGGAHQGEPTGEPESLAGRKLYPMGDKAERG 65
A AF R Y L + P L+ P GA QG + +L L + D RG
Sbjct 307 AAAFRNLSRLRY-----LGVTLSPRLSALPQGAFQGLGELQVLALHSNGLTALPDGLLRG 361
Query 66 LKKEEKEKIGKRRNKLK 82
L K ++ RRN+L+
Sbjct 362 LGK--LRQVSLRRNRLR 376
> hsa:55726 C12orf11, FLJ10630, FLJ10637, NET48; chromosome 12
open reading frame 11
Length=706
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Query 43 PTGEPESLAGRKLYPMGDKAERGLKKEEKEKIGKRRNKLKEGGIAKRAKLDIKKGSTVLD 102
P E GRK DK+E+ +K E+EK + +LK GI +R K ++ + + D
Sbjct 565 PEEEERKKRGRKREDKEDKSEKAVKDYEQEKSWQDSERLK--GILERGKEELAEAEIIKD 622
Query 103 A 103
+
Sbjct 623 S 623
Lambda K H
0.310 0.133 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40