bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8667_orf2
Length=56
Score E
Sequences producing significant alignments: (Bits) Value
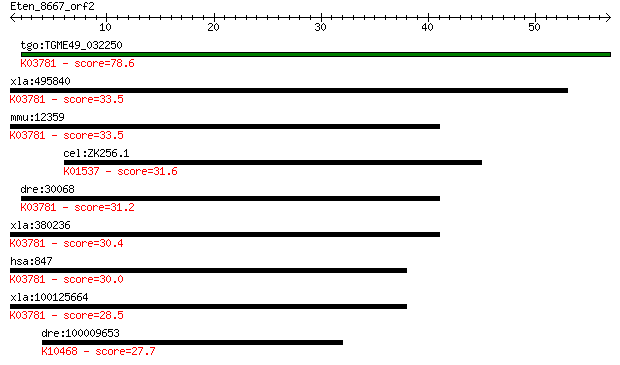
tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781 c... 78.6 5e-15
xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 cata... 33.5 0.17
mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.1... 33.5 0.19
cel:ZK256.1 pmr-1; PMR-type Golgi ATPase family member (pmr-1)... 31.6 0.71
dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6); K0... 31.2 0.74
xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catal... 30.4 1.3
hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03... 30.0 1.6
xla:100125664 hypothetical protein LOC100125664; K03781 catala... 28.5 5.8
dre:100009653 klhl32, zgc:158866; kelch-like 32 (Drosophila); ... 27.7 8.8
> tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=502
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 33/55 (60%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 2 DREALIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRVARGLGLPDSACSSSRM 56
+REALIGNI +HL+ A R +QERQVK+FY+ D +YG RVAR +GLP +AC ++M
Sbjct 448 EREALIGNIAEHLRQARRDIQERQVKIFYKCDPEYGERVARAIGLPTAACYPAKM 502
> xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 DDREALIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRVARGLGLPDSACS 52
++R+ L N+ HLK +Q+R VK F DYG R+ L ++ C+
Sbjct 454 EERQRLCENLAGHLKECQLFIQKRAVKNFSDVHLDYGSRIQALLDKHNAKCT 505
> mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=527
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 1 DDREALIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRV 40
++R+ L NI HLK A +Q++ VK F DYG R+
Sbjct 454 EERKRLCENIAGHLKDAQLFIQKKAVKNFTDVHPDYGARI 493
> cel:ZK256.1 pmr-1; PMR-type Golgi ATPase family member (pmr-1);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=978
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 6 LIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRVARGL 44
L G VD + D L RQV +FYRA + L++ + L
Sbjct 656 LSGQQVDQMSDHDLELVIRQVTVFYRASPRHKLKIVKAL 694
> dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=526
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 2 DREALIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRV 40
+RE L N+ HLK A +Q+R V+ DYG RV
Sbjct 455 ERERLCQNMAGHLKGAQLFIQKRMVQNLMAVHSDYGNRV 493
> xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 1 DDREALIGNIVDHLKHADRHLQERQVKLFYRADKDYGLRV 40
+ R L NI HLK A +Q+R VK F +YG R+
Sbjct 454 EQRLRLCENIAGHLKDAQLFIQKRAVKNFTDVHPEYGARI 493
> hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=527
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 1 DDREALIGNIVDHLKHADRHLQERQVKLFYRADKDYG 37
+ R+ L NI HLK A +Q++ VK F DYG
Sbjct 454 EQRKRLCENIAGHLKDAQIFIQKKAVKNFTEVHPDYG 490
> xla:100125664 hypothetical protein LOC100125664; K03781 catalase
[EC:1.11.1.6]
Length=503
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 1 DDREALIGNIVDHLKHADRHLQERQVKLFYRADKDYG 37
++R+ L N+ +L A +QER VK F DYG
Sbjct 436 EERQRLCENLARNLSEAQIFIQERAVKNFSNIHPDYG 472
> dre:100009653 klhl32, zgc:158866; kelch-like 32 (Drosophila);
K10468 kelch-like protein 32
Length=620
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 4 EALIGNIVDHLKHADRHLQERQVKLFYR 31
EA++G +VDHL R QE + L YR
Sbjct 161 EAVVGFVVDHLSELQRSRQEEVLLLPYR 188
Lambda K H
0.322 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2005549208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40