bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
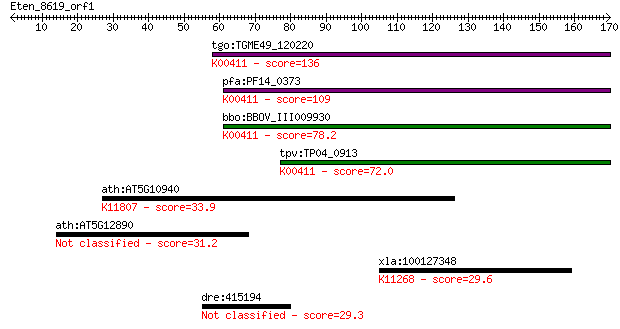
Query= Eten_8619_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_120220 ubiquinol-cytochrome c reductase domain-cont... 136 3e-32
pfa:PF14_0373 ubiquinol-cytochrome c reductase iron-sulfur sub... 109 3e-24
bbo:BBOV_III009930 17.m07861; ubiquinol cytochrome c oxidoredu... 78.2 1e-14
tpv:TP04_0913 ubiquinol-cytochrome C reductase, iron-sulfur su... 72.0 1e-12
ath:AT5G10940 transducin family protein / WD-40 repeat family ... 33.9 0.27
ath:AT5G12890 UDP-glucoronosyl/UDP-glucosyl transferase family... 31.2 1.6
xla:100127348 esco1, eco1, xeco1; establishment of cohesion 1 ... 29.6 5.1
dre:415194 lysmd3, sb:cb462, wu:fb75a04, zgc:86877; LysM, puta... 29.3 7.1
> tgo:TGME49_120220 ubiquinol-cytochrome c reductase domain-containing
protein (EC:1.10.2.2); K00411 ubiquinol-cytochrome
c reductase iron-sulfur subunit [EC:1.10.2.2]
Length=487
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 68/112 (60%), Positives = 82/112 (73%), Gaps = 5/112 (4%)
Query 58 GPMRSFSSFNHNIGPHKSDPPASEKPLYLNRFDQADDPALFQLEAQQLEQQRLLKGQTSD 117
P R FS +HNI P K + PASE PLY NRFDQAD P+L+QLE EQQR K +
Sbjct 154 SPRRHFSVHSHNIRPDKHELPASEVPLYYNRFDQADHPSLWQLEE---EQQR--KHLDQE 208
Query 118 IVDTANVVQPSNHPHQRKGFLKRMRYWHYHQTAEPTFPRTPDLSKGELAAGA 169
+ D + +V+P + PHQ +G+ KR+RYWHY +TAEPTFPRTPDLSKGELAAGA
Sbjct 209 VTDVSQLVEPVSSPHQTEGWFKRLRYWHYKETAEPTFPRTPDLSKGELAAGA 260
> pfa:PF14_0373 ubiquinol-cytochrome c reductase iron-sulfur subunit,
putative; K00411 ubiquinol-cytochrome c reductase iron-sulfur
subunit [EC:1.10.2.2]
Length=355
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 54/109 (49%), Positives = 72/109 (66%), Gaps = 7/109 (6%)
Query 61 RSFSSFNHNIGPHKSDPPASEKPLYLNRFDQADDPALFQLEAQQLEQQRLLKGQTSDIVD 120
R+ +FNHNI ++ PPASE P Y N FD A+D L+++E +Q + ++ D
Sbjct 27 RNGGTFNHNIKENERIPPASEDPSYKNLFDHAEDIKLWEIEEKQNVSHKKVE-------D 79
Query 121 TANVVQPSNHPHQRKGFLKRMRYWHYHQTAEPTFPRTPDLSKGELAAGA 169
+ +V+PSNHPHQ +G R RY HY+QTAEP FPR PDL KGELA+GA
Sbjct 80 LSELVEPSNHPHQYEGIFARTRYAHYNQTAEPVFPRKPDLEKGELASGA 128
> bbo:BBOV_III009930 17.m07861; ubiquinol cytochrome c oxidoreductase
(EC:1.10.2.2); K00411 ubiquinol-cytochrome c reductase
iron-sulfur subunit [EC:1.10.2.2]
Length=332
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 47/112 (41%), Positives = 63/112 (56%), Gaps = 13/112 (11%)
Query 61 RSFSS-FNHNIGPHKSDPPASEKPLYLNRFDQADDPALFQLEAQQLEQQRLLKGQTSDIV 119
RSF+S +H I + P ASE+PLY N FD+A+ PAL+ L+ + E+Q S +
Sbjct 5 RSFASVLHHEIKKTRYMPAASERPLYRNVFDRAEHPALWALDKTRFEKQ-------SSVE 57
Query 120 DTANVVQPSNHPHQ--RKGFLKRMRYWHYHQTAEPTFPRTPDLSKGELAAGA 169
+++ P H H R G K Y HY EP FPRTPD+ KGEL +GA
Sbjct 58 SLGDLITPHAHGHTFARGGITK---YAHYVNVWEPVFPRTPDVMKGELISGA 106
> tpv:TP04_0913 ubiquinol-cytochrome C reductase, iron-sulfur
subunit (EC:1.10.2.2); K00411 ubiquinol-cytochrome c reductase
iron-sulfur subunit [EC:1.10.2.2]
Length=339
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/93 (41%), Positives = 54/93 (58%), Gaps = 8/93 (8%)
Query 77 PPASEKPLYLNRFDQADDPALFQLEAQQLEQQRLLKGQTSDIVDTANVVQPSNHPHQRKG 136
P ASE PLY N FD+AD+P L+ ++ + E+ + + + +++V P H H G
Sbjct 29 PAASEMPLYRNVFDRADNPRLWSMDNTKYEK-------SGPVTELSDLVTPHGHGHL-FG 80
Query 137 FLKRMRYWHYHQTAEPTFPRTPDLSKGELAAGA 169
RY HY EP FPRTPD+SKGEL +GA
Sbjct 81 NTGVTRYAHYVNVWEPVFPRTPDISKGELISGA 113
> ath:AT5G10940 transducin family protein / WD-40 repeat family
protein; K11807 WD and tetratricopeptide repeats protein 1
Length=757
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 50/108 (46%), Gaps = 11/108 (10%)
Query 27 TLTSNGAFPFLKHGGQHSLLRTGNNGVGSVG------GPMRSFSSFNHNIGPHKSDPPAS 80
T + NG L + G+H L NNG+ S G G + + SF++N+ +S P S
Sbjct 286 TFSPNGEEVLLSYSGEHVYLMNVNNGICSTGIMQYTPGDVDNLFSFSNNLHDVESPPQVS 345
Query 81 EKPLYLNRFDQADDPALFQLEAQQLEQQRLLKGQTSDI---VDTANVV 125
P N F ++ + A + + +E + + +D+ ++ AN V
Sbjct 346 TTP--QNGFHRSSNAATVKKCTELVEIAKWSLEEGTDVFYAIEAANEV 391
> ath:AT5G12890 UDP-glucoronosyl/UDP-glucosyl transferase family
protein
Length=488
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 14 RPLLLPRQIGFVSTLTSNGAFPFLKHGGQHSLLRTGNNGVGSVGGPMRSFSSFN 67
R LL+ + V L+ FL H G +S+L + ++GV +G PM + FN
Sbjct 350 RGLLVKKWAPQVDILSHKATCVFLSHCGWNSILESLSHGVPLLGWPMAAEQFFN 403
> xla:100127348 esco1, eco1, xeco1; establishment of cohesion
1 homolog 1; K11268 N-acetyltransferase [EC:2.3.1.-]
Length=967
Score = 29.6 bits (65), Expect = 5.1, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 3/57 (5%)
Query 105 LEQQRLLKGQTSDIVDTANVVQPSNHPHQRKGFLKRMRYWHY---HQTAEPTFPRTP 158
LE+ + K + S IV +A PS P +K +KR + H T+ PT P P
Sbjct 379 LEKSQRPKQKRSSIVASAKKKSPSAKPPHKKLTVKRQKTGHSGTAKNTSAPTLPDVP 435
> dre:415194 lysmd3, sb:cb462, wu:fb75a04, zgc:86877; LysM, putative
peptidoglycan-binding, domain containing 3
Length=305
Score = 29.3 bits (64), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Query 55 SVGGPMRSFSSFN--HNIGPHKSDPPA 79
S+ P+R FSSF HN PHKS P+
Sbjct 109 SLRIPVRKFSSFTETHNTAPHKSSSPS 135
Lambda K H
0.318 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40