bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8595_orf1
Length=149
Score E
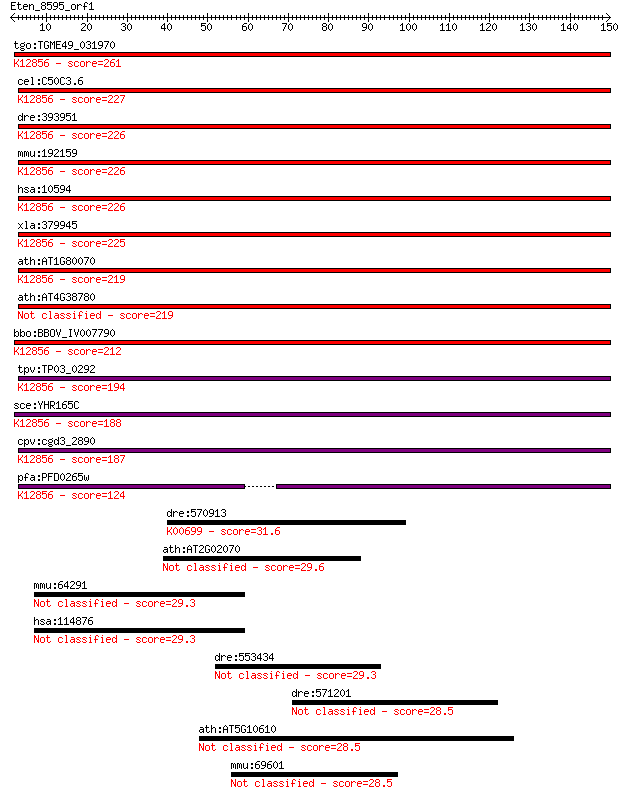
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ; K1... 261 4e-70
cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family ... 227 1e-59
dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2... 226 3e-59
mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfpr... 226 3e-59
hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA proce... 226 3e-59
xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13; P... 225 3e-59
ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mR... 219 2e-57
ath:AT4G38780 splicing factor, putative 219 3e-57
bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12... 212 4e-55
tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing... 194 6e-50
sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component of... 188 6e-48
cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processin... 187 1e-47
pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mR... 124 1e-28
dre:570913 ugt5g1, zgc:175099; UDP glucuronosyltransferase 5 f... 31.6 1.1
ath:AT2G02070 AtIDD5; AtIDD5 (Arabidopsis thaliana Indetermina... 29.6 3.4
mmu:64291 Osbpl1a, G430090F17Rik, Gm753, Osbpl1b; oxysterol bi... 29.3 4.3
hsa:114876 OSBPL1A, FLJ10217, ORP-1, ORP1, OSBPL1B; oxysterol ... 29.3 4.5
dre:553434 msnb, msn; moesin b 29.3 4.8
dre:571201 Kynurenine--oxoglutarate transaminase 3-like 28.5 7.8
ath:AT5G10610 CYP81K1; electron carrier/ heme binding / iron i... 28.5 8.3
mmu:69601 Dab2ip, 2310011D08Rik, AI480459, Aip1, KIAA1743, MGC... 28.5 9.1
> tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ;
K12856 pre-mRNA-processing factor 8
Length=2538
Score = 261 bits (668), Expect = 4e-70, Method: Compositional matrix adjust.
Identities = 122/148 (82%), Positives = 136/148 (91%), Gaps = 0/148 (0%)
Query 2 RDVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQT 61
DVNLGRAVFWE+ NRLPRS++TL W++SFASVYS DNPNLLF+M GFE RILPK+R T
Sbjct 1347 HDVNLGRAVFWEIENRLPRSVSTLEWSNSFASVYSKDNPNLLFAMCGFEVRILPKIRTYT 1406
Query 62 EQFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWN 121
E+F QREGVWKLQNE TKEMAAQAFLKVG++GM+ FENRVR +LM+SGATTFTKIANKWN
Sbjct 1407 EEFSQREGVWKLQNEVTKEMAAQAFLKVGDEGMKHFENRVRQILMASGATTFTKIANKWN 1466
Query 122 TTLISLMTYFREAVVHTEALLDLLVKCE 149
TTLISLMTYFREAV+HTEALLDLLVKCE
Sbjct 1467 TTLISLMTYFREAVIHTEALLDLLVKCE 1494
> cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family
member (prp-8); K12856 pre-mRNA-processing factor 8
Length=2329
Score = 227 bits (578), Expect = 1e-59, Method: Composition-based stats.
Identities = 103/147 (70%), Positives = 120/147 (81%), Gaps = 0/147 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGRAVFW++ NRLPRS+ T+ W +SF SVYS DNPN+LF M GFECRILPK R E
Sbjct 1138 DVNLGRAVFWDIKNRLPRSITTVEWENSFVSVYSKDNPNMLFDMSGFECRILPKCRTANE 1197
Query 63 QFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNT 122
+F+ R+GVW LQNE TKE AQ FLKV E+ + +F NR+R +LMSSG+TTFTKI NKWNT
Sbjct 1198 EFVHRDGVWNLQNEVTKERTAQCFLKVDEESLSKFHNRIRQILMSSGSTTFTKIVNKWNT 1257
Query 123 TLISLMTYFREAVVHTEALLDLLVKCE 149
LI LMTYFREAVV+T+ LLDLLVKCE
Sbjct 1258 ALIGLMTYFREAVVNTQELLDLLVKCE 1284
> dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2a9,
im:7141966, tdsubc_2a9, wu:fb37c02, wu:fb73e06, xx:tdsubc_2a9,
zgc:56504; pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2342
Score = 226 bits (575), Expect = 3e-59, Method: Composition-based stats.
Identities = 103/147 (70%), Positives = 121/147 (82%), Gaps = 0/147 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGRAVFW++ NRLPRS+ T+ W +SF SVYS DNPNLLF+M GFECRILPK R E
Sbjct 1153 DVNLGRAVFWDIKNRLPRSVTTIQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYE 1212
Query 63 QFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNT 122
+F ++GVW LQNE TKE AQ FL+V ++ M+RF NRVR +LM+SG+TTFTKI NKWNT
Sbjct 1213 EFTHKDGVWNLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNT 1272
Query 123 TLISLMTYFREAVVHTEALLDLLVKCE 149
LI LMTYFREAVV+T+ LLDLLVKCE
Sbjct 1273 ALIGLMTYFREAVVNTQELLDLLVKCE 1299
> mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfprp8l;
pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 226 bits (575), Expect = 3e-59, Method: Composition-based stats.
Identities = 103/147 (70%), Positives = 121/147 (82%), Gaps = 0/147 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGRAVFW++ NRLPRS+ T+ W +SF SVYS DNPNLLF+M GFECRILPK R E
Sbjct 1146 DVNLGRAVFWDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYE 1205
Query 63 QFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNT 122
+F ++GVW LQNE TKE AQ FL+V ++ M+RF NRVR +LM+SG+TTFTKI NKWNT
Sbjct 1206 EFTHKDGVWNLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNT 1265
Query 123 TLISLMTYFREAVVHTEALLDLLVKCE 149
LI LMTYFREAVV+T+ LLDLLVKCE
Sbjct 1266 ALIGLMTYFREAVVNTQELLDLLVKCE 1292
> hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA processing
factor 8 homolog (S. cerevisiae); K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 226 bits (575), Expect = 3e-59, Method: Composition-based stats.
Identities = 103/147 (70%), Positives = 121/147 (82%), Gaps = 0/147 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGRAVFW++ NRLPRS+ T+ W +SF SVYS DNPNLLF+M GFECRILPK R E
Sbjct 1146 DVNLGRAVFWDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYE 1205
Query 63 QFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNT 122
+F ++GVW LQNE TKE AQ FL+V ++ M+RF NRVR +LM+SG+TTFTKI NKWNT
Sbjct 1206 EFTHKDGVWNLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNT 1265
Query 123 TLISLMTYFREAVVHTEALLDLLVKCE 149
LI LMTYFREAVV+T+ LLDLLVKCE
Sbjct 1266 ALIGLMTYFREAVVNTQELLDLLVKCE 1292
> xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13;
PRP8 pre-mRNA processing factor 8 homolog; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 225 bits (574), Expect = 3e-59, Method: Composition-based stats.
Identities = 103/147 (70%), Positives = 121/147 (82%), Gaps = 0/147 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGRAVFW++ NRLPRS+ T+ W +SF SVYS DNPNLLF+M GFECRILPK R E
Sbjct 1146 DVNLGRAVFWDIKNRLPRSVTTVQWENSFVSVYSKDNPNLLFNMCGFECRILPKCRTSYE 1205
Query 63 QFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNT 122
+F ++GVW LQNE TKE AQ FL+V ++ M+RF NRVR +LM+SG+TTFTKI NKWNT
Sbjct 1206 EFTHKDGVWNLQNEVTKERTAQCFLRVDDESMQRFHNRVRQILMASGSTTFTKIVNKWNT 1265
Query 123 TLISLMTYFREAVVHTEALLDLLVKCE 149
LI LMTYFREAVV+T+ LLDLLVKCE
Sbjct 1266 ALIGLMTYFREAVVNTQELLDLLVKCE 1292
> ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mRNA-processing
factor 8
Length=2382
Score = 219 bits (559), Expect = 2e-57, Method: Composition-based stats.
Identities = 104/148 (70%), Positives = 119/148 (80%), Gaps = 1/148 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGR+VFW+M NRLPRS+ TL W + F SVYS DNPNLLFSM GFE RILPK+R+ E
Sbjct 1192 DVNLGRSVFWDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRILPKIRMTQE 1251
Query 63 QFLQ-REGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWN 121
F ++GVW LQNE TKE A AFL+V ++ M+ FENRVR +LMSSG+TTFTKI NKWN
Sbjct 1252 AFSNTKDGVWNLQNEQTKERTAVAFLRVDDEHMKVFENRVRQILMSSGSTTFTKIVNKWN 1311
Query 122 TTLISLMTYFREAVVHTEALLDLLVKCE 149
T LI LMTYFREA VHT+ LLDLLVKCE
Sbjct 1312 TALIGLMTYFREATVHTQELLDLLVKCE 1339
> ath:AT4G38780 splicing factor, putative
Length=2332
Score = 219 bits (557), Expect = 3e-57, Method: Composition-based stats.
Identities = 103/148 (69%), Positives = 118/148 (79%), Gaps = 1/148 (0%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQTE 62
DVNLGR+VFW+M NRLPRS+ TL W + F SVYS DNPNLLFSM GFE R+LPK+R+ E
Sbjct 1144 DVNLGRSVFWDMKNRLPRSITTLEWENGFVSVYSKDNPNLLFSMCGFEVRVLPKIRMGQE 1203
Query 63 QFLQ-REGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWN 121
F R+GVW LQNE TKE A AFL+ ++ M+ FENRVR +LMSSG+TTFTKI NKWN
Sbjct 1204 AFSSTRDGVWNLQNEQTKERTAVAFLRADDEHMKVFENRVRQILMSSGSTTFTKIVNKWN 1263
Query 122 TTLISLMTYFREAVVHTEALLDLLVKCE 149
T LI LMTYFREA VHT+ LLDLLVKCE
Sbjct 1264 TALIGLMTYFREATVHTQELLDLLVKCE 1291
> bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12856
pre-mRNA-processing factor 8
Length=2343
Score = 212 bits (539), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 99/148 (66%), Positives = 113/148 (76%), Gaps = 0/148 (0%)
Query 2 RDVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQT 61
DVNLGRA FWEM RLPRS+ TL W SF SVY DNPNLLF+M GFE RI PK+R
Sbjct 1167 HDVNLGRATFWEMQARLPRSVTTLEWNDSFVSVYGKDNPNLLFNMYGFEVRIFPKIRWLK 1226
Query 62 EQFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWN 121
Q E WKLQNE TKE++A A+L+V +GM FENRVR +LM+SG+TTFTKIANKWN
Sbjct 1227 SGVTQAEACWKLQNERTKELSATAYLRVDAEGMSTFENRVRQILMASGSTTFTKIANKWN 1286
Query 122 TTLISLMTYFREAVVHTEALLDLLVKCE 149
T LI +MTY+REAV+HT LLDLLVKCE
Sbjct 1287 TALIGMMTYYREAVIHTNELLDLLVKCE 1314
> tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing
factor 8
Length=2736
Score = 194 bits (494), Expect = 6e-50, Method: Composition-based stats.
Identities = 96/156 (61%), Positives = 114/156 (73%), Gaps = 9/156 (5%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRI--------L 54
DVNLGRA FWEM +RLPRS+ TL W+ SF SVYS DNPNLLFS+ GFE RI +
Sbjct 1565 DVNLGRAAFWEMQSRLPRSITTLEWSDSFVSVYSKDNPNLLFSLCGFEVRIRRYGAANSV 1624
Query 55 PKVRLQTEQFLQ-REGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTF 113
+ T ++ E W+LQN TK+++A A+L+V + M FENRVR +LMSSG+TTF
Sbjct 1625 SDTTVGTVDTVKLSESSWRLQNMKTKQLSAIAYLRVSNESMSMFENRVRQILMSSGSTTF 1684
Query 114 TKIANKWNTTLISLMTYFREAVVHTEALLDLLVKCE 149
TKIANKWNT LISLMTYFREA +HT LLDLLVKCE
Sbjct 1685 TKIANKWNTALISLMTYFREATIHTNELLDLLVKCE 1720
> sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component
of the U4/U6-U5 snRNP complex, involved in the second catalytic
step of splicing; mutations of human Prp8 cause retinitis
pigmentosa; K12856 pre-mRNA-processing factor 8
Length=2413
Score = 188 bits (477), Expect = 6e-48, Method: Composition-based stats.
Identities = 89/148 (60%), Positives = 119/148 (80%), Gaps = 1/148 (0%)
Query 2 RDVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRLQT 61
+DVNLGRAVFWE+ +R+P SL ++ W ++F SVYS +NPNLLFSM GFE RILP+ R++
Sbjct 1218 QDVNLGRAVFWEIQSRVPTSLTSIKWENAFVSVYSKNNPNLLFSMCGFEVRILPRQRME- 1276
Query 62 EQFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWN 121
E EGVW L +E TK+ A+A+LKV E+ +++F++R+R +LM+SG+TTFTK+A KWN
Sbjct 1277 EVVSNDEGVWDLVDERTKQRTAKAYLKVSEEEIKKFDSRIRGILMASGSTTFTKVAAKWN 1336
Query 122 TTLISLMTYFREAVVHTEALLDLLVKCE 149
T+LISL TYFREA+V TE LLD+LVK E
Sbjct 1337 TSLISLFTYFREAIVATEPLLDILVKGE 1364
> cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processing
factor 8
Length=2379
Score = 187 bits (474), Expect = 1e-47, Method: Composition-based stats.
Identities = 89/162 (54%), Positives = 112/162 (69%), Gaps = 15/162 (9%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVRL--- 59
DV +G++V+WE++NRLP+S+ TL W SF SVYS NPNLLFS+ GF RILP R+
Sbjct 1137 DVIIGKSVYWELSNRLPKSITTLEWERSFVSVYSKSNPNLLFSLAGFSVRILPTCRIGKR 1196
Query 60 ------------QTEQFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMS 107
+ Q+ RE W+L N TKE+ + FL V E +R FENRVR +L++
Sbjct 1197 TFEASNTSFIGNEDSQYYTRESTWQLSNNLTKEITSYVFLMVDESEIRNFENRVRQILIT 1256
Query 108 SGATTFTKIANKWNTTLISLMTYFREAVVHTEALLDLLVKCE 149
SG+ TFTKIANKWNT LI LMTYFREAV++TE LLDLLV+CE
Sbjct 1257 SGSATFTKIANKWNTCLIGLMTYFREAVIYTEKLLDLLVRCE 1298
> pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mRNA-processing
factor 8
Length=3136
Score = 124 bits (311), Expect = 1e-28, Method: Composition-based stats.
Identities = 64/83 (77%), Positives = 74/83 (89%), Gaps = 0/83 (0%)
Query 67 REGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGATTFTKIANKWNTTLIS 126
+EG WKLQNE TKE+ A+A+LKV ++ M+RFENRVR +LMSSG+TTFTKIANKWNTTLI
Sbjct 1865 KEGTWKLQNEMTKEITAEAYLKVSDNSMKRFENRVRQILMSSGSTTFTKIANKWNTTLIG 1924
Query 127 LMTYFREAVVHTEALLDLLVKCE 149
LMTYFREAV+ TE LLDLLVKCE
Sbjct 1925 LMTYFREAVLDTEELLDLLVKCE 1947
Score = 90.9 bits (224), Expect = 1e-18, Method: Composition-based stats.
Identities = 40/58 (68%), Positives = 48/58 (82%), Gaps = 2/58 (3%)
Query 3 DVNLGRAVFWEMANRLPRSLATLNWT--SSFASVYSVDNPNLLFSMGGFECRILPKVR 58
DVNLGRA FWE+ NR+PRSL +L+W ++F SVYS DNPNLLFS+ GFE RILPK+R
Sbjct 1693 DVNLGRATFWEIQNRIPRSLTSLDWDHYNTFVSVYSKDNPNLLFSIAGFEVRILPKIR 1750
> dre:570913 ugt5g1, zgc:175099; UDP glucuronosyltransferase 5
family, polypeptide G1; K00699 glucuronosyltransferase [EC:2.4.1.17]
Length=528
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 33/71 (46%), Gaps = 14/71 (19%)
Query 40 PNLLFSMGGFECRILPKVRLQTEQFLQREG------------VWKLQNESTKEMAAQAFL 87
PN+++ MGGF+C+ + + E+F+Q G V L T E A AF
Sbjct 273 PNIIY-MGGFQCKPAQALPVDLEEFMQSSGEHGVVFMSLGAMVGALPRTIT-EAIASAFA 330
Query 88 KVGEDGMRRFE 98
K+ + M R+
Sbjct 331 KIPQKVMWRYH 341
> ath:AT2G02070 AtIDD5; AtIDD5 (Arabidopsis thaliana Indeterminate(ID)-Domain
5); nucleic acid binding / transcription factor/
zinc ion binding
Length=602
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 39 NPNLLFSMGGFECRILPKVRLQTEQFLQ--REG---VWKLQNESTKEMAAQAFL 87
+P L + F C + K Q EQ LQ R G WKL+ +STKE+ + +L
Sbjct 71 SPKTLMATNRFICEVCNK-GFQREQNLQLHRRGHNLPWKLKQKSTKEVKRKVYL 123
> mmu:64291 Osbpl1a, G430090F17Rik, Gm753, Osbpl1b; oxysterol
binding protein-like 1A
Length=950
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 26/52 (50%), Gaps = 1/52 (1%)
Query 7 GRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVR 58
G + W +A R P S N+T SFA V + + + + +CR+ P +R
Sbjct 829 GSVLLWRIAPRPPNSAQMYNFT-SFAMVLNEVDKEMESVIPKTDCRLRPDIR 879
> hsa:114876 OSBPL1A, FLJ10217, ORP-1, ORP1, OSBPL1B; oxysterol
binding protein-like 1A
Length=437
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 7 GRAVFWEMANRLPRSLATLNWTSSFASVYSVDNPNLLFSMGGFECRILPKVR 58
G + W +A R P S N+T SFA V + + ++ + +CR+ P +R
Sbjct 316 GSVLLWRIAPRPPNSAQMYNFT-SFAMVLNEVDKDMESVIPKTDCRLRPDIR 366
> dre:553434 msnb, msn; moesin b
Length=568
Score = 29.3 bits (64), Expect = 4.8, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 52 RILPKVRLQTEQFLQREGVWKLQNES-TKEMAAQAFLKVGED 92
R+L + +L EQ+ QR VW Q++S ++E A +LK+ +D
Sbjct 145 RVLEQHKLTKEQWEQRIQVWHEQHKSMSREDAMIEYLKISQD 186
> dre:571201 Kynurenine--oxoglutarate transaminase 3-like
Length=419
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 71 WKLQNESTKEMAAQAFLKVGEDGMRRFENRVRMVLMSSGAT--TFTKIANKWN 121
W ++ + + AF VGED +++FE +R + +T I KWN
Sbjct 367 WMIKEKKLAAIPVSAF--VGEDSVKQFEKYIRFCFIKQESTLDAAEAILKKWN 417
> ath:AT5G10610 CYP81K1; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=500
Score = 28.5 bits (62), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 19/81 (23%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 48 GFECRILPKVRLQTEQFLQREGVWKLQNESTKEMAAQAFLKVGEDGMRRFENRVR---MV 104
G E R++ R++ E + +++N + + FLK+ E + + V +V
Sbjct 235 GLEKRVIDMQRMRDEYLQRLIDDIRMKNIDSSGSVVEKFLKLQESEPEFYADDVIKGIIV 294
Query 105 LMSSGATTFTKIANKWNTTLI 125
LM +G T + +A +W +L+
Sbjct 295 LMFNGGTDTSPVAMEWAVSLL 315
> mmu:69601 Dab2ip, 2310011D08Rik, AI480459, Aip1, KIAA1743, MGC144147,
mKIAA1743; disabled homolog 2 (Drosophila) interacting
protein
Length=1124
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 56 KVRLQTEQFLQREGVWKLQNESTKEMA--AQAFLKVGEDGMRR 96
K+R+ T++ + E ++K Q E+T+++ QA L+ GE+ +RR
Sbjct 983 KLRISTKKLEEYETLFKCQEETTQKLVLEYQARLEEGEERLRR 1025
Lambda K H
0.322 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40