bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8554_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
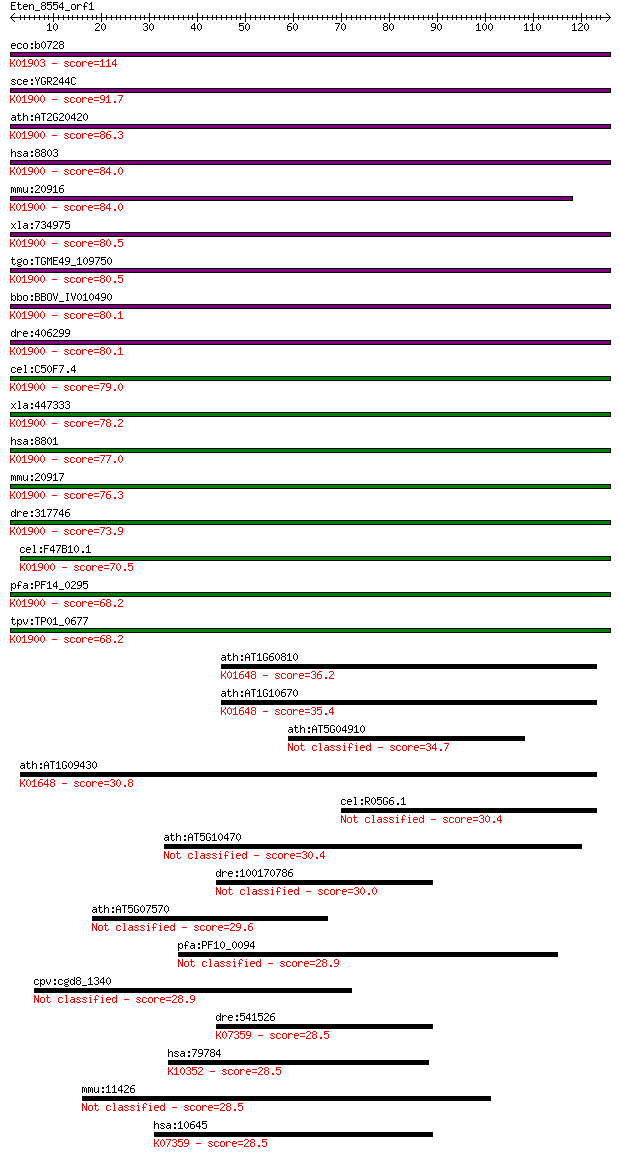
eco:b0728 sucC, ECK0716, JW0717; succinyl-CoA synthetase, beta... 114 8e-26
sce:YGR244C LSC2; Beta subunit of succinyl-CoA ligase, which i... 91.7 6e-19
ath:AT2G20420 succinyl-CoA ligase (GDP-forming) beta-chain, mi... 86.3 2e-17
hsa:8803 SUCLA2, A-BETA, MTDPS5, SCS-betaA; succinate-CoA liga... 84.0 1e-16
mmu:20916 Sucla2, 4930547K18Rik; succinate-Coenzyme A ligase, ... 84.0 1e-16
xla:734975 suclg2, MGC132166, gbeta; succinate-CoA ligase, GDP... 80.5 1e-15
tgo:TGME49_109750 succinyl-CoA ligase, putative (EC:6.2.1.5); ... 80.5 1e-15
bbo:BBOV_IV010490 23.m06275; succinly CoA-ligase beta subunit ... 80.1 2e-15
dre:406299 sucla2, wu:fb18c11, wu:fj38c11, wu:fj90d07, zgc:733... 80.1 2e-15
cel:C50F7.4 hypothetical protein; K01900 succinyl-CoA syntheta... 79.0 4e-15
xla:447333 sucla2, MGC82958; succinate-CoA ligase, ADP-forming... 78.2 5e-15
hsa:8801 SUCLG2, GBETA; succinate-CoA ligase, GDP-forming, bet... 77.0 1e-14
mmu:20917 Suclg2, AF171077, AW556404, D6Wsu120e, MGC91183; suc... 76.3 3e-14
dre:317746 suclg2, cb625, wu:fd44e06; succinate-CoA ligase, GD... 73.9 1e-13
cel:F47B10.1 hypothetical protein; K01900 succinyl-CoA synthet... 70.5 1e-12
pfa:PF14_0295 ATP-specific succinyl-CoA synthetase beta subuni... 68.2 6e-12
tpv:TP01_0677 ATP-specific succinyl-CoA synthetase beta subuni... 68.2 6e-12
ath:AT1G60810 ACLA-2; ACLA-2; ATP citrate synthase (EC:2.3.3.8... 36.2 0.024
ath:AT1G10670 ACLA-1; ACLA-1; ATP citrate synthase (EC:2.3.3.8... 35.4 0.051
ath:AT5G04910 hypothetical protein 34.7 0.070
ath:AT1G09430 ACLA-3; ACLA-3; ATP citrate synthase (EC:2.3.3.8... 30.8 1.1
cel:R05G6.1 hypothetical protein 30.4 1.5
ath:AT5G10470 kinesin motor protein-related 30.4 1.6
dre:100170786 zgc:194737 30.0 2.1
ath:AT5G07570 glycine/proline-rich protein 29.6 2.8
pfa:PF10_0094 tubulin-tyrosine ligase, putative 28.9 3.8
cpv:cgd8_1340 hypothetical protein 28.9 3.9
dre:541526 camkk1, zgc:113440; calcium/calmodulin-dependent pr... 28.5 5.0
hsa:79784 MYH14, DFNA4, DKFZp667A1311, FLJ13881, FLJ43092, KIA... 28.5 6.2
mmu:11426 Macf1, ABP620, Acf7, Aclp7, MACF, R74989, mACF7, mKI... 28.5 6.2
hsa:10645 CAMKK2, CAMKK, CAMKKB, KIAA0787, MGC15254; calcium/c... 28.5 6.3
> eco:b0728 sucC, ECK0716, JW0717; succinyl-CoA synthetase, beta
subunit (EC:6.2.1.5); K01903 succinyl-CoA synthetase beta
subunit [EC:6.2.1.5]
Length=388
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 60/125 (48%), Positives = 80/125 (64%), Gaps = 1/125 (0%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKAGG 60
+N+HEYQAKQ+F +G+ P G + EA A + GP +VVK Q+HAGGRGKAGG
Sbjct 1 MNLHEYQAKQLFARYGLPAPVGYACTTPREAEEAASKIGAGP-WVVKCQVHAGGRGKAGG 59
Query 61 VKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSFTVDRVSS 120
VK+ S ++++ +A LGK LVT QT G+ VN++L+E KELYL VDR S
Sbjct 60 VKVVNSKEDIRAFAENWLGKRLVTYQTDANGQPVNQILVEAATDIAKELYLGAVVDRSSR 119
Query 121 RVVMI 125
RVV +
Sbjct 120 RVVFM 124
> sce:YGR244C LSC2; Beta subunit of succinyl-CoA ligase, which
is a mitochondrial enzyme of the TCA cycle that catalyzes the
nucleotide-dependent conversion of succinyl-CoA to succinate
(EC:6.2.1.4); K01900 succinyl-CoA synthetase beta subunit
[EC:6.2.1.4 6.2.1.5]
Length=427
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 53/132 (40%), Positives = 77/132 (58%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
++IHEY++ Q+ RE+G+ P G PAF+ +EA AK KL V+KAQ GGRGK
Sbjct 31 LSIHEYRSAQLLREYGIGTPEGFPAFTPEEAFEAAK-KLNTNKLVIKAQALTGGRGKGHF 89
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GV + +S + + A E+L L+TKQTG AGK V+ + I + K E YLS
Sbjct 90 DTGYKSGVHMIESPQQAEDVAKEMLNHNLITKQTGIAGKPVSAVYIVKRVDTKHEAYLSI 149
Query 114 TVDRVSSRVVMI 125
+DR + + ++I
Sbjct 150 LMDRQTKKPMII 161
> ath:AT2G20420 succinyl-CoA ligase (GDP-forming) beta-chain,
mitochondrial, putative / succinyl-CoA synthetase, beta chain,
putative / SCS-beta, putative (EC:6.2.1.5 6.2.1.4); K01900
succinyl-CoA synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=421
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 78/133 (58%), Gaps = 9/133 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPV-YVVKAQIHAGGRGKA- 58
+NIHEYQ ++ ++GV VP GV A S++E ++ VVK+QI AGGRG
Sbjct 27 LNIHEYQGAELMGKYGVNVPKGVAASSLEEVKKAIQDVFPNESELVVKSQILAGGRGLGT 86
Query 59 ------GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLS 112
GGV + K DE + A ++LG+VLVTKQTGP GK V+++ + E E+Y S
Sbjct 87 FKSGLKGGVHIVKR-DEAEEIAGKMLGQVLVTKQTGPQGKVVSKVYLCEKLSLVNEMYFS 145
Query 113 FTVDRVSSRVVMI 125
+DR S+ ++I
Sbjct 146 IILDRKSAGPLII 158
> hsa:8803 SUCLA2, A-BETA, MTDPS5, SCS-betaA; succinate-CoA ligase,
ADP-forming, beta subunit (EC:6.2.1.5); K01900 succinyl-CoA
synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=463
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 53/132 (40%), Positives = 81/132 (61%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+++HEY + ++ +E GV+VP G A S DEA A AK KL V+KAQ+ AGGRGK
Sbjct 53 LSLHEYMSMELLQEAGVSVPKGYVAKSPDEAYAIAK-KLGSKDVVIKAQVLAGGRGKGTF 111
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGVK+ S +E + +++++GK L TKQTG G+ N++L+ E ++E Y +
Sbjct 112 ESGLKGGVKIVFSPEEAKAVSSQMIGKKLFTKQTGEKGRICNQVLVCERKYPRREYYFAI 171
Query 114 TVDRVSSRVVMI 125
T++R V+I
Sbjct 172 TMERSFQGPVLI 183
> mmu:20916 Sucla2, 4930547K18Rik; succinate-Coenzyme A ligase,
ADP-forming, beta subunit (EC:6.2.1.5); K01900 succinyl-CoA
synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=463
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 79/124 (63%), Gaps = 8/124 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+++HEY + ++ +E GV+VP G A S DEA A AK KL V+KAQ+ AGGRGK
Sbjct 53 LSLHEYLSMELLQEAGVSVPKGFVAKSSDEAYAIAK-KLGSKDVVIKAQVLAGGRGKGTF 111
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGVK+ S +E + +++++G+ L+TKQTG G+ N++L+ E ++E Y +
Sbjct 112 TSGLKGGVKIVFSPEEAKAVSSQMIGQKLITKQTGEKGRICNQVLVCERKYPRREYYFAI 171
Query 114 TVDR 117
T++R
Sbjct 172 TMER 175
> xla:734975 suclg2, MGC132166, gbeta; succinate-CoA ligase, GDP-forming,
beta subunit; K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=431
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 75/132 (56%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+N+ EYQ+K++ ++GV V + A + EA+ AK KL+ V+KAQI AGGRGK
Sbjct 37 LNLQEYQSKKLMADYGVTVQRFIVADNASEALEAAK-KLKAREIVLKAQILAGGRGKGTF 95
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGV L K +V+ +++G L TKQT G KV ++++ E +E YL+
Sbjct 96 DSGLKGGVHLTKDPSKVEELTKQMIGFNLTTKQTAAGGVKVQKVMVAEALDITRETYLAI 155
Query 114 TVDRVSSRVVMI 125
+DR + V++
Sbjct 156 LMDRACNGPVLV 167
> tgo:TGME49_109750 succinyl-CoA ligase, putative (EC:6.2.1.5);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=498
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 54/141 (38%), Positives = 78/141 (55%), Gaps = 16/141 (11%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKL-EGPV-------YVVKAQIHA 52
+N+HEYQ+ +I +EF + P A + EA A L E P +VVKAQ+ A
Sbjct 72 LNLHEYQSMRIMKEFHITTPKFAVASTAKEAEQEAATFLSESPSGDGEPVDFVVKAQVLA 131
Query 53 GGRGKA--------GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCK 104
GGRG GGV++ +S EV A ++LGK LVTKQTG GK N++L+ E
Sbjct 132 GGRGLGFFRENGYQGGVQVCESPREVGIVAEKMLGKTLVTKQTGKEGKLCNKVLVTERFF 191
Query 105 FKKELYLSFTVDRVSSRVVMI 125
+KE Y++ +DR + ++I
Sbjct 192 IRKEKYVAILMDRGAGGPILI 212
> bbo:BBOV_IV010490 23.m06275; succinly CoA-ligase beta subunit
(EC:6.2.1.5); K01900 succinyl-CoA synthetase beta subunit
[EC:6.2.1.4 6.2.1.5]
Length=436
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/135 (38%), Positives = 72/135 (53%), Gaps = 10/135 (7%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVY---VVKAQIHAGGRGK 57
+N+ E+ +I +E G+ P A S EA A E LE V+KA + GGRGK
Sbjct 39 LNVPEFGGMRILKEHGIPTPMNRLARSPSEAEAMTHEILESTKCGEVVLKALVLTGGRGK 98
Query 58 A-------GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELY 110
GV+LAKS + +T A ++G VLVTKQTG +G K +L+ E KE Y
Sbjct 99 GKFVGTDISGVELAKSPERAKTLAEGMIGNVLVTKQTGASGLKCKEVLVAEKLNLAKERY 158
Query 111 LSFTVDRVSSRVVMI 125
+SF +DR S ++ I
Sbjct 159 ISFMLDRSSCSIMAI 173
> dre:406299 sucla2, wu:fb18c11, wu:fj38c11, wu:fj90d07, zgc:73397;
succinate-CoA ligase, ADP-forming, beta subunit (EC:6.2.1.5);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=466
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 80/132 (60%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+++HEY + + +E G++VP G+ A + DEA AK+ + V+KAQ+ AGGRGK
Sbjct 56 LSLHEYMSIGLLKEAGISVPAGMVASTPDEAYTAAKQ-IGSKDLVIKAQVLAGGRGKGTF 114
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGV++ S +E + +++++GK L TKQTG AG+ N++ + E ++E Y +
Sbjct 115 EGGLKGGVRIVYSPEEARDISSKMIGKKLFTKQTGEAGRICNQVFVCERRYPRREYYFAI 174
Query 114 TVDRVSSRVVMI 125
T++R V+I
Sbjct 175 TMERSYQGPVLI 186
> cel:C50F7.4 hypothetical protein; K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=415
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 54/137 (39%), Positives = 78/137 (56%), Gaps = 16/137 (11%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEG---PVYVVKAQIHAGGRGK 57
+N+ E+Q+K+I + G +V N V A + EA +EK YVVKAQI AGGRGK
Sbjct 20 LNLQEFQSKEILEKHGCSVQNFVVASNRKEA----EEKWMSFGDHEYVVKAQILAGGRGK 75
Query 58 A---------GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKE 108
GGV + K D +E++GK LVTKQT G +V++++I EG K+E
Sbjct 76 GKFINGTKGIGGVFITKEKDAALEAIDEMIGKRLVTKQTTSEGVRVDKVMIAEGVDIKRE 135
Query 109 LYLSFTVDRVSSRVVMI 125
YL+ +DR S+ V++
Sbjct 136 TYLAVLMDRESNGPVVV 152
> xla:447333 sucla2, MGC82958; succinate-CoA ligase, ADP-forming,
beta subunit (EC:6.2.1.5); K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=458
Score = 78.2 bits (191), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 75/132 (56%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+++HEY + + + GVA+P G A + DEA AKE + VVKAQ+ AGGRGK
Sbjct 48 LSLHEYLSMDLLKNAGVAIPKGCVAKTPDEAYTVAKE-IGSKDLVVKAQVLAGGRGKGTF 106
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGVK+ S +E + A++++GK L TKQTG G+ N + I E ++E Y +
Sbjct 107 EGGLKGGVKIVYSPEEAKDIASQMIGKKLFTKQTGEKGRICNHVFICERRYPRREYYFAI 166
Query 114 TVDRVSSRVVMI 125
++R V+I
Sbjct 167 AMERAFQGPVLI 178
> hsa:8801 SUCLG2, GBETA; succinate-CoA ligase, GDP-forming, beta
subunit (EC:6.2.1.4); K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=440
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 49/132 (37%), Positives = 74/132 (56%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+N+ EYQ+K++ + GV V A + +EA+ AK +L V+KAQI AGGRGK
Sbjct 38 LNLQEYQSKKLMSDNGVRVQRFFVADTANEALEAAK-RLNAKEIVLKAQILAGGRGKGVF 96
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGV L K + V A +++G L TKQT G KVN++++ E +E YL+
Sbjct 97 NSGLKGGVHLTKDPNVVGQLAKQMIGYNLATKQTPKEGVKVNKVMVAEALDISRETYLAI 156
Query 114 TVDRVSSRVVMI 125
+DR + V++
Sbjct 157 LMDRSCNGPVLV 168
> mmu:20917 Suclg2, AF171077, AW556404, D6Wsu120e, MGC91183; succinate-Coenzyme
A ligase, GDP-forming, beta subunit (EC:6.2.1.4);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=433
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 72/132 (54%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+N+ EYQ+K++ E GV V A + EA+ AK +L V+KAQI AGGRGK
Sbjct 39 LNLQEYQSKKLMSEHGVRVQRFFVANTAKEALEAAK-RLNAKEIVLKAQILAGGRGKGVF 97
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGV L K V A +++G L TKQT G KVN++++ E +E YL+
Sbjct 98 NSGLKGGVHLTKDPKVVGELAQQMIGYNLATKQTPKEGVKVNKVMVAEALDISRETYLAI 157
Query 114 TVDRVSSRVVMI 125
+DR + V++
Sbjct 158 LMDRSHNGPVIV 169
> dre:317746 suclg2, cb625, wu:fd44e06; succinate-CoA ligase,
GDP-forming, beta subunit; K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=432
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/132 (37%), Positives = 73/132 (55%), Gaps = 8/132 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA-- 58
+N+ EYQ+K++ ++ GVAV A + EA+ AK +L+ V+KAQI AGGRGK
Sbjct 38 LNLQEYQSKKLMQDSGVAVQRFFVADTASEALEAAK-RLKAKEIVLKAQILAGGRGKGVF 96
Query 59 -----GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
GGV L K V A+++LG L TKQT G +V +++ E +E Y +
Sbjct 97 NSGLKGGVHLTKDPAVVGELASKMLGYNLTTKQTPKEGVEVKTVMVAEALDISRETYFAI 156
Query 114 TVDRVSSRVVMI 125
+DR + VM+
Sbjct 157 LMDRSCNGPVMV 168
> cel:F47B10.1 hypothetical protein; K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=435
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 74/130 (56%), Gaps = 8/130 (6%)
Query 3 IHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKA---- 58
+HE+ +I + + + VP A + A + AK ++ G YVVKAQ+ AGGRGK
Sbjct 26 LHEHHGMKILQNYEIKVPPFGVAQDAETAFSEAK-RIGGKDYVVKAQVLAGGRGKGRFSS 84
Query 59 ---GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSFTV 115
GGV++ + DEV+ A ++G L+TKQT GKK +++ + ++E Y S T+
Sbjct 85 GLQGGVQIVFTPDEVKQKAGMMIGANLITKQTDHRGKKCEEVMVCKRLFTRREYYFSITL 144
Query 116 DRVSSRVVMI 125
DR ++ ++I
Sbjct 145 DRNTNGPIVI 154
> pfa:PF14_0295 ATP-specific succinyl-CoA synthetase beta subunit,
putative; K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=462
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 44/142 (30%), Positives = 71/142 (50%), Gaps = 23/142 (16%)
Query 1 VNIHEYQAKQIFREFGVAVPNG---------VPAFSVDEAVAGAKEKLEGPVYVVKAQIH 51
++IHEY + + R V P G + + V G + V+KAQ+
Sbjct 50 LSIHEYLSVDLLRSHNVPCPEGYAAKTAEEAEEKALLLQNVCGDND------LVIKAQVL 103
Query 52 AGGRGKA--------GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGC 103
+GGRG GGV + ++ EV+ A ++L L+TKQ+GP GKK N + I E
Sbjct 104 SGGRGVGYFKENNFEGGVHVCRNSMEVKEIATKMLNNTLITKQSGPEGKKCNTVFICERF 163
Query 104 KFKKELYLSFTVDRVSSRVVMI 125
+KE Y++F +DR S ++++
Sbjct 164 YIRKERYIAFLLDRNSDGIILL 185
> tpv:TP01_0677 ATP-specific succinyl-CoA synthetase beta subunit;
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=453
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 50/134 (37%), Positives = 65/134 (48%), Gaps = 9/134 (6%)
Query 1 VNIHEYQAKQIFREFGVAVPNGVPAFSVDEAVAGAK---EKLEGPVYVVKAQIHAGGRGK 57
+N+ EY I + GV VP A + +EA K + P VVKA + GGRGK
Sbjct 30 LNVSEYIGMTILKRNGVRVPEFRNATTPEEAFEAGKSIQQLTNTPELVVKALVLTGGRGK 89
Query 58 AG------GVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYL 111
GV++ KS EV A +LG L TKQT G +L+ + K E YL
Sbjct 90 GTFNTGFKGVEIVKSPSEVSACARGMLGNYLTTKQTVGKGLLCTEVLVAQKLSIKSERYL 149
Query 112 SFTVDRVSSRVVMI 125
SFT+DR S +V I
Sbjct 150 SFTLDRGSGGIVAI 163
> ath:AT1G60810 ACLA-2; ACLA-2; ATP citrate synthase (EC:2.3.3.8);
K01648 ATP citrate (pro-S)-lyase [EC:2.3.3.8]
Length=423
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 45 VVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGK-VLVTKQTGPAGKKVNRLLIEEGC 103
VVK + G RGK+G V L +V T+ E LGK V ++ GP + ++E
Sbjct 56 VVKPDMLFGKRGKSGLVALNLDFADVATFVKERLGKEVEMSGCKGP----ITTFIVEPFV 111
Query 104 KFKKELYLSFTVDRVSSRV 122
+E YL+ DR+ +
Sbjct 112 PHNEEFYLNIVSDRLGCSI 130
> ath:AT1G10670 ACLA-1; ACLA-1; ATP citrate synthase (EC:2.3.3.8);
K01648 ATP citrate (pro-S)-lyase [EC:2.3.3.8]
Length=423
Score = 35.4 bits (80), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 45 VVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGK-VLVTKQTGPAGKKVNRLLIEEGC 103
VVK + G RGK+G V L +V T+ E LGK V ++ GP + ++E
Sbjct 56 VVKPDMLFGKRGKSGLVALKLDFADVATFVKERLGKEVEMSGCKGP----ITTFIVEPFV 111
Query 104 KFKKELYLSFTVDRVSSRV 122
+E YL+ DR+ +
Sbjct 112 PHNEEYYLNVVSDRLGCSI 130
> ath:AT5G04910 hypothetical protein
Length=222
Score = 34.7 bits (78), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 59 GGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKK 107
G LAK L ++QT + L + + AG+ R+L E CKFKK
Sbjct 112 GRADLAKLLSDIQTQEKQKLHLTVTIQVLKKAGRPSERMLTHENCKFKK 160
> ath:AT1G09430 ACLA-3; ACLA-3; ATP citrate synthase (EC:2.3.3.8);
K01648 ATP citrate (pro-S)-lyase [EC:2.3.3.8]
Length=424
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 33/129 (25%), Positives = 54/129 (41%), Gaps = 13/129 (10%)
Query 3 IHEYQAKQIFREFGVAVPN---GVPAFSVDEA-----VAGAKEKLEGPVYVVKAQIHAGG 54
I EY +K++ +E + N + + V E+ + + L VVK + G
Sbjct 6 IREYDSKRLLKEHLKRLANIDLQIRSAQVTESTDFTELTNQESWLSSTKLVVKPDMLFGK 65
Query 55 RGKAGGVKLAKSLDEVQTYANEILG-KVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSF 113
RGK+G V L L EV + LG +V + P + ++E +E YLS
Sbjct 66 RGKSGLVALKLDLAEVADFVKARLGTEVEMEGCKAP----ITTFIVEPFVPHDQEYYLSI 121
Query 114 TVDRVSSRV 122
DR+ +
Sbjct 122 VSDRLGCTI 130
> cel:R05G6.1 hypothetical protein
Length=314
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Query 70 VQTYANEILGKVLVTKQTGPAGKKVNRLLIEEGCKFKKELYLSFTVDRVSSRV 122
+QT +E+ G ++T+ +G ++ L + GC F KE + T D++ + V
Sbjct 211 IQTNTDELKGARVMTQTSG-----LSSLFLNCGCAFSKETWSVLTQDKIMANV 258
> ath:AT5G10470 kinesin motor protein-related
Length=1274
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 27/89 (30%), Positives = 42/89 (47%), Gaps = 8/89 (8%)
Query 33 AGAKEKLEGPVYVVKAQIHAGGRGKAGGVKLAKSLDEVQTYA--NEILGKVLVTKQTGPA 90
A + KL V ++ GR + GVKL + DE +++A N+ L + V G
Sbjct 864 AERRNKLASVVSRMRGLEQDAGRQQVTGVKLREMQDEAKSFAIGNKALAALFVHTPAGEL 923
Query 91 GKKVNRLLIEEGCKFKKELYLSFTVDRVS 119
+++ RL + E +F LS T D VS
Sbjct 924 QRQI-RLWLAENFEF-----LSVTSDDVS 946
> dre:100170786 zgc:194737
Length=450
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 3/45 (6%)
Query 44 YVVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGKVLVTKQTG 88
Y +K +I G+G G VKLA + D+ Q YA +++ K + KQ G
Sbjct 93 YKLKNEI---GKGSYGVVKLAYNEDDDQYYAMKLVSKKRLIKQLG 134
> ath:AT5G07570 glycine/proline-rich protein
Length=1504
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 18 AVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKAGGVKLAKS 66
A P G+ S+D VAGA + EGP+ + + G+G +G L +S
Sbjct 651 AGPLGISPLSIDPPVAGASD--EGPLGKLPPDLSPPGKGSSGEGPLVES 697
> pfa:PF10_0094 tubulin-tyrosine ligase, putative
Length=553
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 16/79 (20%)
Query 36 KEKLEGPVYVVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGKVLVTKQTGPAGKKVN 95
K K Y+VK + G+G + L KSLD++ Y + I+ K ++
Sbjct 141 KRKGSSKTYIVKLKNSCQGKG----IYLTKSLDDINKYESCIIQKY------------IH 184
Query 96 RLLIEEGCKFKKELYLSFT 114
+ L+ G KF LY+ T
Sbjct 185 KPLLINGLKFDIRLYVLLT 203
> cpv:cgd8_1340 hypothetical protein
Length=2806
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 29/71 (40%), Gaps = 5/71 (7%)
Query 6 YQAKQIFREFGV-----AVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGGRGKAGG 60
Y+ K I EF + ++ +P + + G +L P VK +HA G
Sbjct 765 YKGKVILAEFDINLTCASMDQHLPYSGAKDTLGGKSSELHAPDLQVKNHLHADNESDKGH 824
Query 61 VKLAKSLDEVQ 71
V LDE++
Sbjct 825 VLKYVGLDEIE 835
> dre:541526 camkk1, zgc:113440; calcium/calmodulin-dependent
protein kinase kinase 1, alpha (EC:2.7.11.17); K07359 calcium/calmodulin-dependent
protein kinase kinase [EC:2.7.11.17]
Length=434
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 28/45 (62%), Gaps = 3/45 (6%)
Query 44 YVVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGKVLVTKQTG 88
Y +K++I G+G G VKLA + D+ + YA +++ K + KQ G
Sbjct 56 YKLKSEI---GKGSYGVVKLAYNEDDDKYYAMKVVSKKKLMKQYG 97
> hsa:79784 MYH14, DFNA4, DKFZp667A1311, FLJ13881, FLJ43092, KIAA2034,
MHC16, MYH17, NMHC-II-C, myosin; myosin, heavy chain
14, non-muscle (EC:3.6.4.1); K10352 myosin heavy chain
Length=2003
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 34 GAKEKLEGPVYVVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGKVLVTKQT 87
A++KLEG + +KAQ+ + G+GK V K L ++Q E+ +V T+ +
Sbjct 1642 AARKKLEGELEELKAQMASAGQGKEEAV---KQLRKMQAQMKELWREVEETRTS 1692
> mmu:11426 Macf1, ABP620, Acf7, Aclp7, MACF, R74989, mACF7, mKIAA0465;
microtubule-actin crosslinking factor 1
Length=7355
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 39/91 (42%), Gaps = 7/91 (7%)
Query 16 GVAVPNGVPAFSVDEAVAGAKEKLEGPVYVVKAQIHAGG-----RGKAGGVKLAKSLDEV 70
G+ P SV EAVA E +++ Q+ GG RGK V LA +L V
Sbjct 2362 GILDPRTHSLCSVKEAVAAGLLDKETATRILEGQVITGGIVDLKRGKKLSVTLASNLGLV 2421
Query 71 QTY-ANEILGKVLVTKQTGPAGKKVNRLLIE 100
T E++ TK G A K V LIE
Sbjct 2422 DTADQTELINLEKATKGRG-AEKAVKERLIE 2451
> hsa:10645 CAMKK2, CAMKK, CAMKKB, KIAA0787, MGC15254; calcium/calmodulin-dependent
protein kinase kinase 2, beta (EC:2.7.11.17);
K07359 calcium/calmodulin-dependent protein kinase kinase
[EC:2.7.11.17]
Length=588
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Query 31 AVAGAKEKLEGPVYVVKAQIHAGGRGKAGGVKLAKSLDEVQTYANEILGKVLVTKQTG 88
++ G ++ ++ Y +K +I G+G G VKLA + ++ YA ++L K + +Q G
Sbjct 152 SITGMQDCVQLNQYTLKDEI---GKGSYGVVKLAYNENDNTYYAMKVLSKKKLIRQAG 206
Lambda K H
0.317 0.135 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40