bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8450_orf1
Length=163
Score E
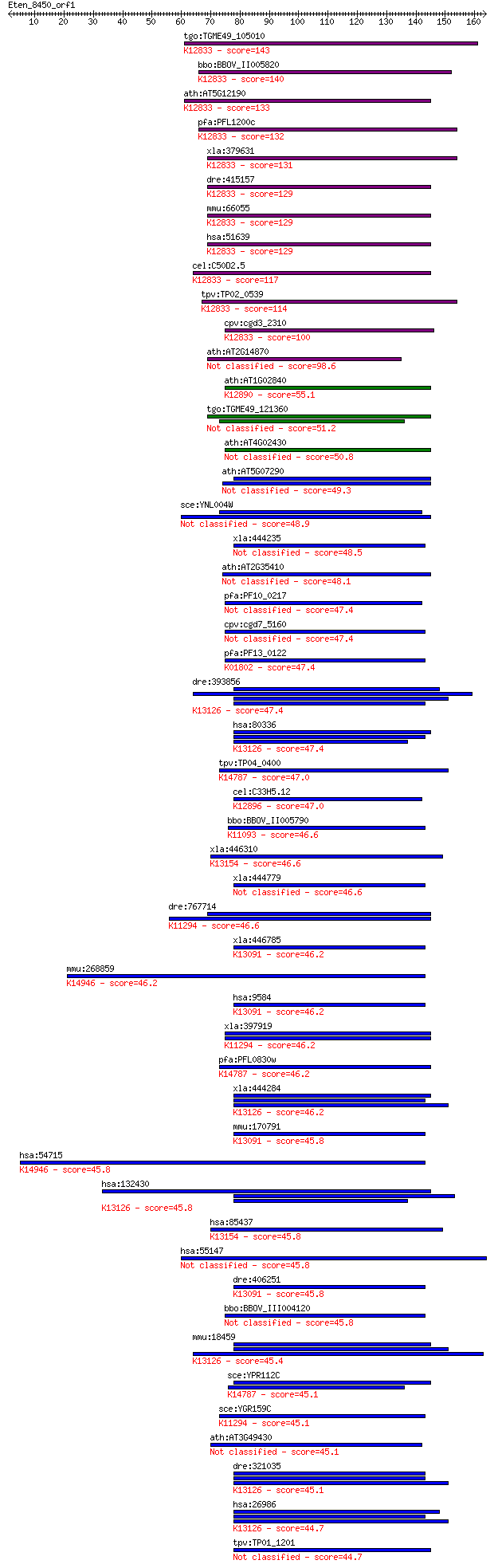
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_105010 RNA binding protein, putative ; K12833 pre-m... 143 3e-34
bbo:BBOV_II005820 18.m06483; pre-mRNA branch site protein p14;... 140 1e-33
ath:AT5G12190 RNA recognition motif (RRM)-containing protein; ... 133 3e-31
pfa:PFL1200c splicing factor 3b subunit, putative; K12833 pre-... 132 7e-31
xla:379631 sf3b14, MGC68842; splicing factor 3B, 14 kDa subuni... 131 1e-30
dre:415157 sf3b14, zgc:86708; splicing factor 3B; K12833 pre-m... 129 4e-30
mmu:66055 0610009D07Rik, 6030419K15Rik, AV001342, Sf3b14; RIKE... 129 4e-30
hsa:51639 SF3B14, HSPC175, Ht006, P14, SAP14, SF3B14a; splicin... 129 4e-30
cel:C50D2.5 hypothetical protein; K12833 pre-mRNA branch site ... 117 1e-26
tpv:TP02_0539 hypothetical protein; K12833 pre-mRNA branch sit... 114 1e-25
cpv:cgd3_2310 hypothetical protein ; K12833 pre-mRNA branch si... 100 3e-21
ath:AT2G14870 RNA recognition motif (RRM)-containing protein 98.6 7e-21
ath:AT1G02840 SR1; SR1; RNA binding / nucleic acid binding / n... 55.1 1e-07
tgo:TGME49_121360 RNA recognition motif domain-containing protein 51.2 1e-06
ath:AT4G02430 pre-mRNA splicing factor, putative / SR1 protein... 50.8 2e-06
ath:AT5G07290 AML4; AML4 (ARABIDOPSIS MEI2-LIKE 4); RNA bindin... 49.3 5e-06
sce:YNL004W HRB1, TOM34; Hrb1p 48.9 8e-06
xla:444235 rbm23, MGC80803; RNA binding motif protein 23 48.5
ath:AT2G35410 33 kDa ribonucleoprotein, chloroplast, putative ... 48.1 1e-05
pfa:PF10_0217 pre-mRNA splicing factor, putative 47.4 2e-05
cpv:cgd7_5160 U1 small nuclear ribonucleoprotein A, rrm domain 47.4 2e-05
pfa:PF13_0122 RRM containing cyclophilin (EC:5.2.1.8); K01802 ... 47.4 2e-05
dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A bindin... 47.4 2e-05
hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053, PA... 47.4 2e-05
tpv:TP04_0400 hypothetical protein; K14787 multiple RNA-bindin... 47.0 2e-05
cel:C33H5.12 rsp-6; SR Protein (splicing factor) family member... 47.0 3e-05
bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear... 46.6 3e-05
xla:446310 zcrb1, MGC82154; zinc finger CCHC-type and RNA bind... 46.6 4e-05
xla:444779 MGC81970 protein 46.6 4e-05
dre:767714 ncl, MGC152810, wu:fa12d03, zgc:152810; nucleolin; ... 46.6 4e-05
xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein 3... 46.2 4e-05
mmu:268859 Rbfox1, A2bp, A2bp1, Hrnbp1, fox-1; RNA binding pro... 46.2 5e-05
hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44... 46.2 5e-05
xla:397919 ncl; nucleolin; K11294 nucleolin 46.2 5e-05
pfa:PFL0830w RNA binding protein, putative; K14787 multiple RN... 46.2 5e-05
xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) bindin... 46.2 5e-05
mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,... 45.8 6e-05
hsa:54715 RBFOX1, A2BP1, FOX-1, FOX1, HRNBP1; RNA binding prot... 45.8 6e-05
hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein, c... 45.8 6e-05
hsa:85437 ZCRB1, MADP-1, MADP1, MGC26805, RBM36, ZCCHC19; zinc... 45.8 6e-05
hsa:55147 RBM23, CAPERbeta, FLJ10482, MGC4458, RNPC4; RNA bind... 45.8 6e-05
dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein... 45.8 6e-05
bbo:BBOV_III004120 17.m07378; U2 small nuclear ribonucleoprote... 45.8 7e-05
mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,... 45.4 9e-05
sce:YPR112C MRD1; Essential conserved protein that is part of ... 45.1 1e-04
sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin 45.1 1e-04
ath:AT3G49430 SRp34a; SRp34a (Ser/Arg-rich protein 34a); RNA b... 45.1 1e-04
dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cyt... 45.1 1e-04
hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A) b... 44.7 1e-04
tpv:TP01_1201 hypothetical protein 44.7 1e-04
> tgo:TGME49_105010 RNA binding protein, putative ; K12833 pre-mRNA
branch site protein p14
Length=156
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 62/100 (62%), Positives = 81/100 (81%), Gaps = 0/100 (0%)
Query 61 LAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVV 120
++ RGRP ++AP++SRI+YVRNLPFKI +ELYD+FGKYG++RQIR+G + TRG+AFVV
Sbjct 37 VSTRGRPTKIAPDMSRIIYVRNLPFKITDDELYDIFGKYGAVRQIRKGNTDKTRGSAFVV 96
Query 121 YDDIYDAKAAVDQLSGFNVAGRYLGGAVLQPHKGAAAERC 160
Y+D+ DA+AAVDQLSGFNVAGRYL P K A ++
Sbjct 97 YEDVLDARAAVDQLSGFNVAGRYLIVLYYNPQKAAQRQQM 136
> bbo:BBOV_II005820 18.m06483; pre-mRNA branch site protein p14;
K12833 pre-mRNA branch site protein p14
Length=122
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 62/86 (72%), Positives = 71/86 (82%), Gaps = 0/86 (0%)
Query 66 RPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIY 125
R RL+PEVSRILY+RNLP+KI+ EELYD+FGKYGS+RQIR+G + T GTAFVVYDDIY
Sbjct 10 RTMRLSPEVSRILYLRNLPYKISAEELYDIFGKYGSVRQIRKGNTSKTNGTAFVVYDDIY 69
Query 126 DAKAAVDQLSGFNVAGRYLGGAVLQP 151
DAK A+D LSGFNVAGRYL P
Sbjct 70 DAKNALDHLSGFNVAGRYLVVLYYNP 95
> ath:AT5G12190 RNA recognition motif (RRM)-containing protein;
K12833 pre-mRNA branch site protein p14
Length=124
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 59/84 (70%), Positives = 68/84 (80%), Gaps = 0/84 (0%)
Query 61 LAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVV 120
++ R RL PEV+R+LYVRNLPF I EE+YD+FGKYG+IRQIR G AT+GTAFVV
Sbjct 4 ISLRKSNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGCDKATKGTAFVV 63
Query 121 YDDIYDAKAAVDQLSGFNVAGRYL 144
Y+DIYDAK AVD LSGFNVA RYL
Sbjct 64 YEDIYDAKNAVDHLSGFNVANRYL 87
> pfa:PFL1200c splicing factor 3b subunit, putative; K12833 pre-mRNA
branch site protein p14
Length=106
Score = 132 bits (331), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 60/88 (68%), Positives = 71/88 (80%), Gaps = 0/88 (0%)
Query 66 RPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIY 125
R RL EVSRILYVRNLP+KI+ +ELYD+FGKYG++RQIR+G + T+GT+FVVYDDIY
Sbjct 4 RNIRLPAEVSRILYVRNLPYKISADELYDIFGKYGTVRQIRKGNAEGTKGTSFVVYDDIY 63
Query 126 DAKAAVDQLSGFNVAGRYLGGAVLQPHK 153
DAK A+D LSGFNVAGRYL P K
Sbjct 64 DAKNALDHLSGFNVAGRYLVVLYYDPVK 91
> xla:379631 sf3b14, MGC68842; splicing factor 3B, 14 kDa subunit;
K12833 pre-mRNA branch site protein p14
Length=125
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 56/85 (65%), Positives = 68/85 (80%), Gaps = 0/85 (0%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
RL PEV+RILY+RNLP+KI GEE+YD+FGKYG IRQIR G +P +RGTA+VVY+DI+DAK
Sbjct 12 RLPPEVNRILYIRNLPYKITGEEMYDIFGKYGPIRQIRVGNTPESRGTAYVVYEDIFDAK 71
Query 129 AAVDQLSGFNVAGRYLGGAVLQPHK 153
A D LSGFNV RYL P++
Sbjct 72 NACDHLSGFNVCNRYLVVLYYNPNR 96
> dre:415157 sf3b14, zgc:86708; splicing factor 3B; K12833 pre-mRNA
branch site protein p14
Length=125
Score = 129 bits (324), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 55/76 (72%), Positives = 64/76 (84%), Gaps = 0/76 (0%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
RL PEV+RILY+RNLP+KI EE+YD+FGKYG IRQIR G +P TRGTA+VVY+DI+DAK
Sbjct 12 RLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTAYVVYEDIFDAK 71
Query 129 AAVDQLSGFNVAGRYL 144
A D LSGFNV RYL
Sbjct 72 NACDHLSGFNVCNRYL 87
> mmu:66055 0610009D07Rik, 6030419K15Rik, AV001342, Sf3b14; RIKEN
cDNA 0610009D07 gene; K12833 pre-mRNA branch site protein
p14
Length=125
Score = 129 bits (324), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 55/76 (72%), Positives = 64/76 (84%), Gaps = 0/76 (0%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
RL PEV+RILY+RNLP+KI EE+YD+FGKYG IRQIR G +P TRGTA+VVY+DI+DAK
Sbjct 12 RLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTAYVVYEDIFDAK 71
Query 129 AAVDQLSGFNVAGRYL 144
A D LSGFNV RYL
Sbjct 72 NACDHLSGFNVCNRYL 87
> hsa:51639 SF3B14, HSPC175, Ht006, P14, SAP14, SF3B14a; splicing
factor 3B, 14 kDa subunit; K12833 pre-mRNA branch site protein
p14
Length=125
Score = 129 bits (324), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 55/76 (72%), Positives = 64/76 (84%), Gaps = 0/76 (0%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
RL PEV+RILY+RNLP+KI EE+YD+FGKYG IRQIR G +P TRGTA+VVY+DI+DAK
Sbjct 12 RLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTAYVVYEDIFDAK 71
Query 129 AAVDQLSGFNVAGRYL 144
A D LSGFNV RYL
Sbjct 72 NACDHLSGFNVCNRYL 87
> cel:C50D2.5 hypothetical protein; K12833 pre-mRNA branch site
protein p14
Length=138
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 49/81 (60%), Positives = 67/81 (82%), Gaps = 0/81 (0%)
Query 64 RGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDD 123
+ R +L PEV+RILY++NLP+KI EE+Y++FGK+G++RQIR G + TRGTAFVVY+D
Sbjct 7 QNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGNTAETRGTAFVVYED 66
Query 124 IYDAKAAVDQLSGFNVAGRYL 144
I+DAK A + LSG+NV+ RYL
Sbjct 67 IFDAKTACEHLSGYNVSNRYL 87
> tpv:TP02_0539 hypothetical protein; K12833 pre-mRNA branch site
protein p14
Length=134
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 56/97 (57%), Positives = 65/97 (67%), Gaps = 10/97 (10%)
Query 67 PQRLAPEVSRILYVR----------NLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGT 116
P EV R + +R NLP+KI EELYD+FGKYGS+RQIR+G S T+GT
Sbjct 13 PTTFGLEVQRAIQLRHGGSSKTSNLNLPYKITSEELYDIFGKYGSVRQIRKGNSATTKGT 72
Query 117 AFVVYDDIYDAKAAVDQLSGFNVAGRYLGGAVLQPHK 153
AFVVYDDI+DAK A+D LSGFNVAGRYL P K
Sbjct 73 AFVVYDDIFDAKNALDHLSGFNVAGRYLVVLYYNPAK 109
> cpv:cgd3_2310 hypothetical protein ; K12833 pre-mRNA branch
site protein p14
Length=86
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 43/71 (60%), Positives = 58/71 (81%), Gaps = 0/71 (0%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
S I+Y+R LP+ I+ +LYD+FG++G+IRQIRRG T+GTAFVVYD+I DAK+A+ QL
Sbjct 15 SSIIYLRQLPYDISSTDLYDIFGRHGTIRQIRRGVGEDTKGTAFVVYDEIEDAKSALKQL 74
Query 135 SGFNVAGRYLG 145
SGF V+GRY+
Sbjct 75 SGFQVSGRYVN 85
> ath:AT2G14870 RNA recognition motif (RRM)-containing protein
Length=101
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
RL PEV+R+LY+ NLPF I E+ YD+FG+Y +IRQ+R G T+GTAFVVY+DIYDAK
Sbjct 13 RLPPEVTRLLYICNLPFSITSEDTYDLFGRYSTIRQVRIGCEKGTKGTAFVVYEDIYDAK 72
Query 129 AAVDQL 134
AVD L
Sbjct 73 KAVDHL 78
> ath:AT1G02840 SR1; SR1; RNA binding / nucleic acid binding /
nucleotide binding; K12890 splicing factor, arginine/serine-rich
1/9
Length=303
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
SR +YV NLP I E+ D+F KYG + QI P G AFV +DD DA+ A+
Sbjct 6 SRTVYVGNLPGDIREREVEDLFSKYGPVVQIDLKVPPRPPGYAFVEFDDARDAEDAIHGR 65
Query 135 SGFNVAGRYL 144
G++ G L
Sbjct 66 DGYDFDGHRL 75
> tgo:TGME49_121360 RNA recognition motif domain-containing protein
Length=274
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIR--QIRRGTSPATRGTAFVVYDDIYD 126
R P S + +V NLP++ N +L F +G+I +I+RGT +RG FV +D+
Sbjct 161 RFGPPGSNV-FVANLPYEWNDIDLIQHFQHFGNILSARIQRGTEGNSRGFGFVSFDNSQA 219
Query 127 AKAAVDQLSGFNVAGRYL 144
A A+ ++GF+ GRYL
Sbjct 220 AVNAIRGMNGFSCGGRYL 237
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 38/65 (58%), Gaps = 2/65 (3%)
Query 73 EVSRILYVRNLPFKINGEELYDVFGKYGSIRQI--RRGTSPATRGTAFVVYDDIYDAKAA 130
+V++ ++V ++P + EE ++GS+ + + + RG AFV Y+ +YDA+ A
Sbjct 3 KVTKKIFVGSIPHTVTEEEFRKKVEEHGSVTALFYMKDQTEGDRGWAFVTYETVYDAQNA 62
Query 131 VDQLS 135
++ L+
Sbjct 63 IEALN 67
> ath:AT4G02430 pre-mRNA splicing factor, putative / SR1 protein,
putative
Length=278
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
SR +YV NLP I E+ D+F KYG + QI P G AFV ++D DA A+
Sbjct 6 SRTIYVGNLPGDIREREVEDLFSKYGPVVQIDLKIPPRPPGYAFVEFEDARDADDAIYGR 65
Query 135 SGFNVAGRYL 144
G++ G +L
Sbjct 66 DGYDFDGHHL 75
> ath:AT5G07290 AML4; AML4 (ARABIDOPSIS MEI2-LIKE 4); RNA binding
/ nucleic acid binding / nucleotide binding
Length=907
Score = 49.3 bits (116), Expect = 5e-06, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 40/67 (59%), Gaps = 2/67 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQLSGF 137
L+V NL I+ EEL+ +F YG IR++RR ++ ++ + D+ AK A+ L+G
Sbjct 297 LWVNNLDSSISNEELHGIFSSYGEIREVRRTMHENSQ--VYIEFFDVRKAKVALQGLNGL 354
Query 138 NVAGRYL 144
VAGR L
Sbjct 355 EVAGRQL 361
Score = 39.3 bits (90), Expect = 0.005, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 2/71 (2%)
Query 74 VSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQ 133
+SRIL+VRN+ I EL +F ++G +R + T+ RG V Y DI A+ A
Sbjct 209 LSRILFVRNVDSSIEDCELGVLFKQFGDVRALH--TAGKNRGFIMVSYYDIRAAQKAARA 266
Query 134 LSGFNVAGRYL 144
L G + GR L
Sbjct 267 LHGRLLRGRKL 277
> sce:YNL004W HRB1, TOM34; Hrb1p
Length=454
Score = 48.9 bits (115), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 2/71 (2%)
Query 73 EVSRILYVRNLPFKINGEELYDVFGKYGSIR--QIRRGTSPATRGTAFVVYDDIYDAKAA 130
E +R++Y NLPF +LYD+F G + ++R + A G A V YD++ DA
Sbjct 373 ERNRLIYCSNLPFSTAKSDLYDLFETIGKVNNAELRYDSKGAPTGIAVVEYDNVDDADVC 432
Query 131 VDQLSGFNVAG 141
+++L+ +N G
Sbjct 433 IERLNNYNYGG 443
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 42/87 (48%), Gaps = 6/87 (6%)
Query 60 LLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQ--IRRGTSPATRGTA 117
L G R R EV V+NLP +N + L D+F + G++ + + G+
Sbjct 249 LDRGELRHNRKTHEV----IVKNLPASVNWQALKDIFKECGNVAHADVELDGDGVSTGSG 304
Query 118 FVVYDDIYDAKAAVDQLSGFNVAGRYL 144
V + DI D A+++ +G+++ G L
Sbjct 305 TVSFYDIKDLHRAIEKYNGYSIEGNVL 331
> xla:444235 rbm23, MGC80803; RNA binding motif protein 23
Length=416
Score = 48.5 bits (114), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I E L +F +G I I+ P T +G F+ + D A+ A++QL
Sbjct 252 LYVGSLHFNITEEMLRGIFEPFGKIENIQLLKEPDTGRSKGFGFITFTDAECARRALEQL 311
Query 135 SGFNVAGR 142
+GF +AG+
Sbjct 312 NGFELAGK 319
> ath:AT2G35410 33 kDa ribonucleoprotein, chloroplast, putative
/ RNA-binding protein cp33, putative
Length=308
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 43/73 (58%), Gaps = 2/73 (2%)
Query 74 VSRILYVRNLPFKINGEELYDVFGKYGSIR--QIRRGTSPATRGTAFVVYDDIYDAKAAV 131
+ R L+V NLP+ ++ ++ ++FG+ G++ +I R RG AFV +A+AA+
Sbjct 93 LKRKLFVFNLPWSMSVNDISELFGQCGTVNNVEIIRQKDGKNRGFAFVTMASGEEAQAAI 152
Query 132 DQLSGFNVAGRYL 144
D+ F V+GR +
Sbjct 153 DKFDTFQVSGRII 165
> pfa:PF10_0217 pre-mRNA splicing factor, putative
Length=538
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/68 (39%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIR-RGTSPATRGTAFVVYDDIYDAKAAVDQ 133
S +YV NLP + EE+YD+FGKYG I+ I + + ++ AFV Y D+ DA A+++
Sbjct 11 SSCIYVGNLPGNVIEEEVYDLFGKYGRIKYIDIKPSRSSSSSYAFVHYYDLKDADYAIER 70
Query 134 LSGFNVAG 141
G+ G
Sbjct 71 RDGYKFDG 78
> cpv:cgd7_5160 U1 small nuclear ribonucleoprotein A, rrm domain
Length=122
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 46/73 (63%), Gaps = 6/73 (8%)
Query 75 SRILYVRNLPFKINGEEL----YDVFGKYGSIRQIR-RGTSPATRGTAFVVYDDIYDAKA 129
++ LY +NL KIN ++L +++F ++G I +I RG SP RG AF+++ D+ A
Sbjct 16 NKTLYCKNLNDKINKKDLKILLFELFIQFGLIDKITIRGGSPY-RGQAFILFQDLNCAIN 74
Query 130 AVDQLSGFNVAGR 142
A + ++G NV G+
Sbjct 75 AKNSINGMNVLGK 87
> pfa:PF13_0122 RRM containing cyclophilin (EC:5.2.1.8); K01802
peptidylprolyl isomerase [EC:5.2.1.8]
Length=125
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIR---RGTSPATRGTAFVVYDDIYDAKAAV 131
+ IL+V + I+ + LYD+F +G IR I T+ RG AFV Y ++ DAK A+
Sbjct 7 TDILFVGGIDETIDEKSLYDIFSSFGDIRNIEVPLNMTTKKNRGFAFVEYVEVDDAKHAL 66
Query 132 DQLSGFNVAGR 142
++ F + G+
Sbjct 67 YNMNNFELNGK 77
> dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A binding
protein, cytoplasmic 1 b; K13126 polyadenylate-binding protein
Length=634
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 44/74 (59%), Gaps = 4/74 (5%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP--ATRGTAFVVYDDIYDAKAAVDQLS 135
+Y++N ++ ++L D+F KYG+ IR T +RG FV ++ DA+ AVD+++
Sbjct 193 VYIKNFGEDMDDDKLKDIFSKYGNAMSIRVMTDENGKSRGFGFVSFERHEDAQRAVDEMN 252
Query 136 GFNVAGR--YLGGA 147
G + G+ Y+G A
Sbjct 253 GKEMNGKLIYVGRA 266
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 24/105 (22%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Query 64 RGRPQRLA-----PEVSR----ILYVRNLPFKINGEELYDVFGKYGSIRQIR-RGTSPAT 113
+GRP R+ P + + ++++NL I+ + LYD F +G+I + +
Sbjct 78 KGRPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGS 137
Query 114 RGTAFVVYDDIYDAKAAVDQLSGFNVAGRYLGGAVLQPHKGAAAE 158
+G FV ++ A+ A+++++G + R + + K AE
Sbjct 138 KGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAE 182
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 37/74 (50%), Gaps = 1/74 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA-TRGTAFVVYDDIYDAKAAVDQLSG 136
LYV+NL I+ E L F +G+I + ++G FV + +A AV +++G
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMDGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query 137 FNVAGRYLGGAVLQ 150
VA + L A+ Q
Sbjct 356 RIVATKPLYVALAQ 369
Score = 28.9 bits (63), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATR---GTAFVVYDDIYDAKAAVDQL 134
LYV +L + LY+ F G+I IR TR G A+V + DA+ A+D +
Sbjct 13 LYVGDLHQDVTEAMLYEKFSPAGAILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query 135 SGFNVAGR 142
+ + GR
Sbjct 73 NFDVIKGR 80
> hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053,
PABPC1L1, dJ1069P2.3, ePAB; poly(A) binding protein, cytoplasmic
1-like; K13126 polyadenylate-binding protein
Length=614
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 41/69 (59%), Gaps = 2/69 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIR--RGTSPATRGTAFVVYDDIYDAKAAVDQLS 135
+YV+NLP ++ + L D+F ++G + ++ R S +R FV ++ +A+ AV ++
Sbjct 193 IYVKNLPVDVDEQGLQDLFSQFGKMLSVKVMRDNSGHSRCFGFVNFEKHEEAQKAVVHMN 252
Query 136 GFNVAGRYL 144
G V+GR L
Sbjct 253 GKEVSGRLL 261
Score = 36.2 bits (82), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP-ATRGTAFVVYDDIYDAKAAVDQLSG 136
++++NL I+ + LYD F +G+I + +RG FV ++ A+ A++ ++G
Sbjct 101 IFIKNLEDSIDNKALYDTFSTFGNILSCKVACDEHGSRGFGFVHFETHEAAQQAINTMNG 160
Query 137 FNVAGR 142
+ R
Sbjct 161 MLLNDR 166
Score = 32.0 bits (71), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA-TRGTAFVVYDDIYDAKAAVDQLSG 136
LYV+NL I+ ++L F YG I + T ++G FV + +A AV +++G
Sbjct 296 LYVKNLDDSIDDDKLRKEFSPYGVITSAKVMTEGGHSKGFGFVCFSSPEEATKAVTEMNG 355
> tpv:TP04_0400 hypothetical protein; K14787 multiple RNA-binding
domain-containing protein 1
Length=727
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 45/80 (56%), Gaps = 5/80 (6%)
Query 73 EVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR--RGTSPATRGTAFVVYDDIYDAKAA 130
E + ++ V+NLPF+ +EL ++F Y +++ +R + RG FVV+ DAK A
Sbjct 645 EENDVIIVKNLPFQATKKELLELFKYYSNVKTVRIPKSAGNTHRGFGFVVFMSKNDAKLA 704
Query 131 VDQLSGFNVAGRYLGGAVLQ 150
++ L ++ GR L VLQ
Sbjct 705 MENLKNVHLYGRRL---VLQ 721
> cel:C33H5.12 rsp-6; SR Protein (splicing factor) family member
(rsp-6); K12896 splicing factor, arginine/serine-rich 7
Length=179
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 36/64 (56%), Gaps = 2/64 (3%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQLSGF 137
+YV LP +EL ++F ++G IR++ P G AFV YDD+ DA+ AV L G
Sbjct 5 VYVGGLPSDATSQELEEIFDRFGRIRKVWVARRPP--GFAFVEYDDVRDAEDAVRALDGS 62
Query 138 NVAG 141
+ G
Sbjct 63 RICG 66
> bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear
ribonucleoprotein 70kDa
Length=247
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Query 76 RILYVRNLPFKINGEELYDVFGKYGSIRQIR----RGTSPATRGTAFVVYDDIYDAKAAV 131
+ L+V N+P+++ ++L+ F YG +R+IR R P RG AF+ Y+D D A
Sbjct 107 KTLFVSNIPYEVTEKQLWKEFDVYGRVRRIRMINDRKNIP--RGYAFIEYNDERDMVNAY 164
Query 132 DQLSGFNVAGR 142
+ G V+GR
Sbjct 165 KRADGRKVSGR 175
> xla:446310 zcrb1, MGC82154; zinc finger CCHC-type and RNA binding
motif 1; K13154 U11/U12 small nuclear ribonucleoprotein
31 kDa protein
Length=218
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Query 70 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI---RRGTSPATRGTAFVVYDDIYD 126
LAP S + YV NLPF + +L+ +F KYG + ++ + S ++G +FV++ D
Sbjct 5 LAPSKSTV-YVSNLPFSLTNNDLHRIFSKYGKVVKVTILKDKDSRKSKGVSFVLFLDKES 63
Query 127 AKAAVDQLSGFNVAGRYLGGAV 148
A+ V L+ + GR + ++
Sbjct 64 AQNCVRGLNNKQLFGRAIKASI 85
> xla:444779 MGC81970 protein
Length=512
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I + L +F +G I I+ T +G F+ + D AK A++QL
Sbjct 225 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 284
Query 135 SGFNVAGR 142
+GF +AGR
Sbjct 285 NGFELAGR 292
> dre:767714 ncl, MGC152810, wu:fa12d03, zgc:152810; nucleolin;
K11294 nucleolin
Length=705
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 43/76 (56%), Gaps = 2/76 (2%)
Query 69 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAK 128
R AP S++L V NL F + + L VF K SIR + P +G AFV ++++ D+K
Sbjct 458 RGAPSASKVLVVNNLAFSASEDSLQSVFEKAVSIRIPQNNGRP--KGYAFVEFENVEDSK 515
Query 129 AAVDQLSGFNVAGRYL 144
A++ + ++ GR +
Sbjct 516 EALENCNNTDIEGRSI 531
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 48/92 (52%), Gaps = 4/92 (4%)
Query 56 QPAPLLAGRGRP---QRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA 112
QP L R + + +R L+V+NLP+ I ++L ++F + IR + G +
Sbjct 354 QPVKLDKARSKENSQENKKERDARTLFVKNLPYSITQDDLREIFDQAVDIR-VPMGNTGT 412
Query 113 TRGTAFVVYDDIYDAKAAVDQLSGFNVAGRYL 144
+RG A++ + A+ A+++ G +V GR +
Sbjct 413 SRGIAYIEFKTEAIAEKALEEAQGSDVQGRSI 444
> xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein
39; K13091 RNA-binding protein 39
Length=540
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I + L +F +G I I+ T +G F+ + D AK A++QL
Sbjct 251 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 310
Query 135 SGFNVAGR 142
+GF +AGR
Sbjct 311 NGFELAGR 318
> mmu:268859 Rbfox1, A2bp, A2bp1, Hrnbp1, fox-1; RNA binding protein,
fox-1 homolog (C. elegans) 1; K14946 RNA binding protein
fox-1
Length=396
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 59/127 (46%), Gaps = 21/127 (16%)
Query 21 AEAAASLSASFAAIAQTPTESAAAAAAAAAKSPAMQPAPLLAGRGRPQRLAPEVSRILYV 80
+E +A SA + T T+ AA P QP+ + +P+RL +V
Sbjct 73 SEQSADTSAQTVSGTATQTDDAAPTDG----QPQTQPSENTESKSQPKRL--------HV 120
Query 81 RNLPFKINGEELYDVFGKYGSIRQI-----RRGTSPATRGTAFVVYDDIYDAKAAVDQLS 135
N+PF+ +L +FG++G I + RG ++G FV +++ DA A ++L
Sbjct 121 SNIPFRFRDPDLRQMFGQFGKILDVEIIFNERG----SKGFGFVTFENSADADRAREKLH 176
Query 136 GFNVAGR 142
G V GR
Sbjct 177 GTVVEGR 183
> hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44170,
HCC1, RNPC2, fSAP59; RNA binding motif protein 39; K13091
RNA-binding protein 39
Length=524
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I + L +F +G I I+ T +G F+ + D AK A++QL
Sbjct 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query 135 SGFNVAGR 142
+GF +AGR
Sbjct 312 NGFELAGR 319
> xla:397919 ncl; nucleolin; K11294 nucleolin
Length=705
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
SR L+V+N+P+ + EEL ++F IR I G + +G A+V + +A A+++
Sbjct 375 SRTLFVKNIPYSTSAEELQEIFENAKDIR-IPTGNDGSNKGIAYVEFSTEAEANKALEEK 433
Query 135 SGFNVAGRYL 144
G + GR L
Sbjct 434 QGAEIEGRSL 443
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Query 75 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
S++L V NL + + L +VF K SIR I + A +G AFV + + DAK A+D
Sbjct 465 SKVLVVNNLSYSATEDSLREVFEKATSIR-IPQNQGRA-KGFAFVEFSSMEDAKEAMDSC 522
Query 135 SGFNVAGRYL 144
+ V GR +
Sbjct 523 NNTEVEGRSI 532
> pfa:PFL0830w RNA binding protein, putative; K14787 multiple
RNA-binding domain-containing protein 1
Length=1119
Score = 46.2 bits (108), Expect = 5e-05, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 45/74 (60%), Gaps = 2/74 (2%)
Query 73 EVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR--RGTSPATRGTAFVVYDDIYDAKAA 130
+V++ L V+NL F++N EEL +F +G+++ +R + +RG AF+ + ++ A
Sbjct 980 QVTKKLVVKNLAFQVNKEELRKLFSAFGNVKSVRIPKNVYNRSRGYAFIEFMSKKESCNA 1039
Query 131 VDQLSGFNVAGRYL 144
++ L ++ GR+L
Sbjct 1040 IESLQHTHLYGRHL 1053
> xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) binding
protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=626
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 39/69 (56%), Gaps = 2/69 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA--TRGTAFVVYDDIYDAKAAVDQLS 135
+Y++N ++ E L + F KYG ++ T P+ ++G FV ++ DA AVD ++
Sbjct 193 VYIKNFGEDMDDERLKETFSKYGKTLSVKVMTDPSGKSKGFGFVSFERHEDANKAVDDMN 252
Query 136 GFNVAGRYL 144
G +V G+ +
Sbjct 253 GKDVNGKIM 261
Score = 33.1 bits (74), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 36/66 (54%), Gaps = 1/66 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP-ATRGTAFVVYDDIYDAKAAVDQLSG 136
++++NL I+ + LYD F +G+I + ++G AFV ++ A A+++++G
Sbjct 101 VFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYAFVHFETQDAADRAIEKMNG 160
Query 137 FNVAGR 142
+ R
Sbjct 161 MLLNDR 166
Score = 32.7 bits (73), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIR----QIRRGTSPATRGTAFVVYDDIYDAKAAVDQ 133
LY++NL I+ E+L F +GSI + G S +G FV + +A AV +
Sbjct 296 LYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLEEGRS---KGFGFVCFSSPEEATKAVTE 352
Query 134 LSGFNVAGRYLGGAVLQ 150
++G V + L A+ Q
Sbjct 353 MNGRIVGSKPLYVALAQ 369
> mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,
C79248, R75070, Rnpc2, caper; RNA binding motif protein 39;
K13091 RNA-binding protein 39
Length=530
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I + L +F +G I I+ T +G F+ + D AK A++QL
Sbjct 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query 135 SGFNVAGR 142
+GF +AGR
Sbjct 312 NGFELAGR 319
> hsa:54715 RBFOX1, A2BP1, FOX-1, FOX1, HRNBP1; RNA binding protein,
fox-1 homolog (C. elegans) 1; K14946 RNA binding protein
fox-1
Length=370
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 62/143 (43%), Gaps = 19/143 (13%)
Query 5 TTAARATAAAAAAAGTAEAAASLSASFAAIAQTPTESAAAAAAAAAKSPAMQPAPLLAGR 64
TT T A T + S ++ T T++ AA P QP+ +
Sbjct 56 TTVPEHTLNLYPPAQTHSEQSPADTSAQTVSGTATQTDDAAPTDG--QPQTQPSENTENK 113
Query 65 GRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI-----RRGTSPATRGTAFV 119
+P+RL +V N+PF+ +L +FG++G I + RG ++G FV
Sbjct 114 SQPKRL--------HVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERG----SKGFGFV 161
Query 120 VYDDIYDAKAAVDQLSGFNVAGR 142
+++ DA A ++L G V GR
Sbjct 162 TFENSADADRAREKLHGTVVEGR 184
> hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein,
cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=428
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 56/121 (46%), Gaps = 9/121 (7%)
Query 33 AIAQTPTESAAAAAAAAAKSPAMQPAPLLAGR--GRPQRLAPEVSR-----ILYVRNLPF 85
A +SAA A ++ + GR R R A S+ +Y++N
Sbjct 198 AFVHFQNQSAADRAIEEMNGKLLKGCKVFVGRFKNRKDREAELRSKASEFTNVYIKNFGG 257
Query 86 KINGEELYDVFGKYGSIRQIRRGT--SPATRGTAFVVYDDIYDAKAAVDQLSGFNVAGRY 143
++ E L DVF KYG ++ T S ++G FV +D AK AV++++G ++ G+
Sbjct 258 DMDDERLKDVFSKYGKTLSVKVMTDSSGKSKGFGFVSFDSHEAAKKAVEEMNGRDINGQL 317
Query 144 L 144
+
Sbjct 318 I 318
Score = 34.7 bits (78), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA-TRGTAFVVYDDIYDAKAAVDQLSG 136
LY++NL I+ E+L + F +GSI +++ ++G + + DA A+ +++G
Sbjct 353 LYIKNLDDTIDDEKLRNEFSSFGSISRVKVMQEEGQSKGFGLICFSSPEDATKAMTEMNG 412
Query 137 FNVAGRYLGGAVLQPH 152
+ + L A+ Q H
Sbjct 413 RILGSKPLSIALAQRH 428
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIR-RGTSPATRGTAFVVYDDIYDAKAAVDQLSG 136
++++NL I+ + LY+ F +G I + ++G AFV + + A A+++++G
Sbjct 158 VFIKNLDKSIDNKTLYEHFSAFGKILSSKVMSDDQGSKGYAFVHFQNQSAADRAIEEMNG 217
> hsa:85437 ZCRB1, MADP-1, MADP1, MGC26805, RBM36, ZCCHC19; zinc
finger CCHC-type and RNA binding motif 1; K13154 U11/U12
small nuclear ribonucleoprotein 31 kDa protein
Length=217
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 42/82 (51%), Gaps = 4/82 (4%)
Query 70 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATR---GTAFVVYDDIYD 126
LAP S + YV NLPF + +LY +F KYG + ++ TR G AF+++ D
Sbjct 5 LAPSKSTV-YVSNLPFSLTNNDLYRIFSKYGKVVKVTIMKDKDTRKSKGVAFILFLDKDS 63
Query 127 AKAAVDQLSGFNVAGRYLGGAV 148
A+ ++ + GR + ++
Sbjct 64 AQNCTRAINNKQLFGRVIKASI 85
> hsa:55147 RBM23, CAPERbeta, FLJ10482, MGC4458, RNPC4; RNA binding
motif protein 23
Length=439
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 51/107 (47%), Gaps = 18/107 (16%)
Query 60 LLAGRGRPQRLAPEVSRILYVRNLPFKINGEEL---YDVFGKYGSIRQIRRGTSPATRGT 116
L G G P RL YV +L F I + L ++ FGK +I ++ + ++G
Sbjct 255 LQKGNGGPMRL--------YVGSLHFNITEDMLRGIFEPFGKIDNIVLMKDSDTGRSKGY 306
Query 117 AFVVYDDIYDAKAAVDQLSGFNVAGRYLGGAVLQPHKGAAAERCAGG 163
F+ + D A+ A++QL+GF +AGR + G ER GG
Sbjct 307 GFITFSDSECARRALEQLNGFELAGRPM-------RVGHVTERLDGG 346
> dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein
39a; K13091 RNA-binding protein 39
Length=523
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 3/68 (4%)
Query 78 LYVRNLPFKINGEEL---YDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
LYV +L F I + L ++ FG+ SI+ + + ++G F+ + D AK A++QL
Sbjct 248 LYVGSLHFNITEDMLRGIFEPFGRIDSIQLMMDSETGRSKGYGFITFSDAECAKKALEQL 307
Query 135 SGFNVAGR 142
+GF +AGR
Sbjct 308 NGFELAGR 315
> bbo:BBOV_III004120 17.m07378; U2 small nuclear ribonucleoprotein
B'
Length=285
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 42/72 (58%), Gaps = 4/72 (5%)
Query 75 SRILYVRNLPFKINGE----ELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAA 130
++ +Y++N+ +I + EL +FG++G I I TS +G AF+++D++ A A
Sbjct 30 NQTIYIKNINERIKVDVLKAELLKMFGRFGKILDIVALTSFWRKGQAFIIFDNVESATNA 89
Query 131 VDQLSGFNVAGR 142
+ ++ GF + G
Sbjct 90 LHEMQGFVMDGH 101
> mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,
cytoplasmic 2; K13126 polyadenylate-binding protein
Length=628
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 42/69 (60%), Gaps = 2/69 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP--ATRGTAFVVYDDIYDAKAAVDQLS 135
+Y++N +++ E L +FG++G I ++ T ++G FV ++ DA+ AVD+++
Sbjct 193 VYIKNFGDRMDDETLNGLFGRFGQILSVKVMTDEGGKSKGFGFVSFERHEDAQKAVDEMN 252
Query 136 GFNVAGRYL 144
G + G+++
Sbjct 253 GKELNGKHI 261
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 38/74 (51%), Gaps = 1/74 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA-TRGTAFVVYDDIYDAKAAVDQLSG 136
LYV+NL I+ E L F +G+I + T ++G FV + +A AV +++G
Sbjct 296 LYVKNLDDGIDDERLQKEFSPFGTITSTKVMTEGGRSKGFGFVCFSSPEEATKAVSEMNG 355
Query 137 FNVAGRYLGGAVLQ 150
VA + L A+ Q
Sbjct 356 RIVATKPLYVALAQ 369
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 25/109 (22%), Positives = 52/109 (47%), Gaps = 10/109 (9%)
Query 64 RGRPQRLA-----PEVSR----ILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP-AT 113
+G+P R+ P + R ++++NL I+ + LYD F +G+I + + +
Sbjct 78 KGKPVRIMWSQRDPSLRRSGVGNVFIKNLNKTIDNKALYDTFSAFGNILSCKVVSDENGS 137
Query 114 RGTAFVVYDDIYDAKAAVDQLSGFNVAGRYLGGAVLQPHKGAAAERCAG 162
+G FV ++ A+ A+++++G + R + + K AE G
Sbjct 138 KGHGFVHFETEEAAERAIEKMNGMLLNDRKVFVGRFKSQKEREAELGTG 186
> sce:YPR112C MRD1; Essential conserved protein that is part of
the 90S preribosome; required for production of 18S rRNA and
small ribosomal subunit; contains five consensus RNA-binding
domains; K14787 multiple RNA-binding domain-containing protein
1
Length=887
Score = 45.1 bits (105), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 43/69 (62%), Gaps = 2/69 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIR--RGTSPATRGTAFVVYDDIYDAKAAVDQLS 135
+ V+NLPF+ ++++++F +G ++ +R + + RG AFV + +A+ A+DQL
Sbjct 765 IIVKNLPFEATRKDVFELFNSFGQLKSVRVPKKFDKSARGFAFVEFLLPKEAENAMDQLH 824
Query 136 GFNVAGRYL 144
G ++ GR L
Sbjct 825 GVHLLGRRL 833
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 76 RILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKAAVDQLS 135
+++ V+N PF EEL ++F YG ++ R P A V + D A+AA +LS
Sbjct 532 KVILVKNFPFGTTREELGEMFLPYG---KLERLLMPPAGTIAIVQFRDTTSARAAFTKLS 588
> sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin
Length=414
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 41/73 (56%), Gaps = 3/73 (4%)
Query 73 EVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPAT---RGTAFVVYDDIYDAKA 129
E S L++ NL F + + ++++F K+G + +R T P T +G +V + ++ DAK
Sbjct 264 EPSDTLFLGNLSFNADRDAIFELFAKHGEVVSVRIPTHPETEQPKGFGYVQFSNMEDAKK 323
Query 130 AVDQLSGFNVAGR 142
A+D L G + R
Sbjct 324 ALDALQGEYIDNR 336
> ath:AT3G49430 SRp34a; SRp34a (Ser/Arg-rich protein 34a); RNA
binding / nucleic acid binding / nucleotide binding
Length=297
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 70 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPATRGTAFVVYDDIYDAKA 129
++ SR +YV NLP I E+ D+F KYG I I P FV ++ DA+
Sbjct 1 MSGRFSRSIYVGNLPGDIREHEIEDIFYKYGRIVDIELKVPPRPPCYCFVEFEHSRDAED 60
Query 130 AVDQLSGFNVAG 141
A+ G+N+ G
Sbjct 61 AIKGRDGYNLDG 72
> dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cytoplasmic
4 (inducible form); K13126 polyadenylate-binding
protein
Length=637
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 38/67 (56%), Gaps = 2/67 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSPA--TRGTAFVVYDDIYDAKAAVDQLS 135
+Y++N ++ + L ++F KYG ++ T P +RG FV Y+ DA AV++++
Sbjct 194 VYIKNFGDDMDDQRLKELFDKYGKTLSVKVMTDPTGKSRGFGFVSYEKHEDANKAVEEMN 253
Query 136 GFNVAGR 142
G + G+
Sbjct 254 GTELNGK 260
Score = 33.5 bits (75), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 36/66 (54%), Gaps = 1/66 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIR-RGTSPATRGTAFVVYDDIYDAKAAVDQLSG 136
++++NL I+ + LYD F +G+I + ++G AFV ++ A A+++++G
Sbjct 102 VFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYAFVHFETQDAADRAIEKMNG 161
Query 137 FNVAGR 142
+ R
Sbjct 162 MLLNDR 167
Score = 32.7 bits (73), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIR----QIRRGTSPATRGTAFVVYDDIYDAKAAVDQ 133
LY++NL I+ E+L F +GSI + G S +G FV + +A AV +
Sbjct 297 LYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLEEGRS---KGFGFVCFSSPEEATKAVTE 353
Query 134 LSGFNVAGRYLGGAVLQ 150
++G V + L A+ Q
Sbjct 354 MNGRIVGSKPLYVALAQ 370
> hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 44/74 (59%), Gaps = 4/74 (5%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGT--SPATRGTAFVVYDDIYDAKAAVDQLS 135
+Y++N ++ E L D+FGK+G ++ T S ++G FV ++ DA+ AVD+++
Sbjct 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query 136 GFNVAGR--YLGGA 147
G + G+ Y+G A
Sbjct 253 GKELNGKQIYVGRA 266
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 1/66 (1%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGTSP-ATRGTAFVVYDDIYDAKAAVDQLSG 136
++++NL I+ + LYD F +G+I + ++G FV ++ A+ A+++++G
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNG 160
Query 137 FNVAGR 142
+ R
Sbjct 161 MLLNDR 166
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 5/76 (6%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSI---RQIRRGTSPATRGTAFVVYDDIYDAKAAVDQL 134
LYV+NL I+ E L F +G+I + + G ++G FV + +A AV ++
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGR--SKGFGFVCFSSPEEATKAVTEM 353
Query 135 SGFNVAGRYLGGAVLQ 150
+G VA + L A+ Q
Sbjct 354 NGRIVATKPLYVALAQ 369
> tpv:TP01_1201 hypothetical protein
Length=268
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 41/69 (59%), Gaps = 2/69 (2%)
Query 78 LYVRNLPFKINGEELYDVFGKYGSIR--QIRRGTSPATRGTAFVVYDDIYDAKAAVDQLS 135
L+V ++P N +L + F +G++ +++R ++ RG F+ YD+ A A+ ++
Sbjct 156 LFVFHVPANWNDLDLVEHFKHFGNVISARVQRDSAGRNRGFGFISYDNPQSAVVAIKNMN 215
Query 136 GFNVAGRYL 144
GF+V G+YL
Sbjct 216 GFSVGGKYL 224
Lambda K H
0.315 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3832864436
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40