bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8437_orf1
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
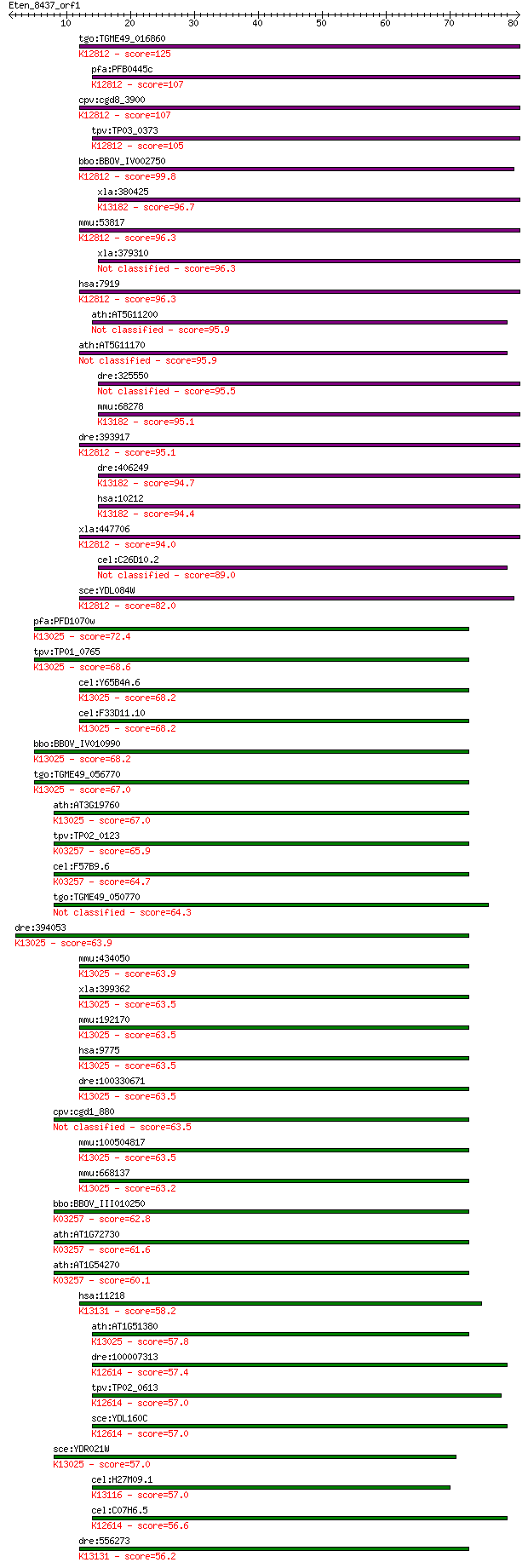
tgo:TGME49_016860 ATP-dependent RNA helicase, putative ; K1281... 125 3e-29
pfa:PFB0445c UAP56, U52; DEAD box helicase, UAP56; K12812 ATP-... 107 8e-24
cpv:cgd8_3900 Sub2p like superfamily II helicase involved in s... 107 9e-24
tpv:TP03_0373 ATP-dependent RNA helicase; K12812 ATP-dependent... 105 5e-23
bbo:BBOV_IV002750 21.m02887; eIF-4A-like DEAD family RNA helic... 99.8 2e-21
xla:380425 ddx39, MGC53944; nuclear RNA helicase; K13182 ATP-d... 96.7 2e-20
mmu:53817 Ddx39b, 0610030D10Rik, AI428441, Bat-1, Bat1, Bat1a,... 96.3 2e-20
xla:379310 ddx39a, MGC130793, MGC53693, bat1, bat1l, ddx39, dd... 96.3 2e-20
hsa:7919 DDX39B, BAT1, D6S81E, UAP56; DEAD (Asp-Glu-Ala-Asp) b... 96.3 2e-20
ath:AT5G11200 DEAD/DEAH box helicase, putative 95.9 2e-20
ath:AT5G11170 ATP binding / ATP-dependent helicase/ helicase/ ... 95.9 3e-20
dre:325550 ddx39a, wu:fc87b12, zgc:55881; DEAD (Asp-Glu-Ala-As... 95.5 4e-20
mmu:68278 Ddx39, 2610307C23Rik, BAT1, DDXL, Ddx39a, URH49; DEA... 95.1 5e-20
dre:393917 bat1, Bat1a, MGC63773, zgc:63773; HLA-B associated ... 95.1 5e-20
dre:406249 ddx39b, ddx39, wu:fc16a02, zgc:55433, zgc:85646; DE... 94.7 7e-20
hsa:10212 DDX39A, BAT1, BAT1L, DDX39, DDXL, MGC18203, MGC8417,... 94.4 9e-20
xla:447706 ddx39b, MGC81606, bat1, uap56; DEAD (Asp-Glu-Ala-As... 94.0 1e-19
cel:C26D10.2 hel-1; HELicase family member (hel-1) 89.0 4e-18
sce:YDL084W SUB2; Component of the TREX complex required for n... 82.0 5e-16
pfa:PFD1070w eukaryotic initiation factor, putative; K13025 AT... 72.4 3e-13
tpv:TP01_0765 eukaryotic translation initiation factor 4A; K13... 68.6 4e-12
cel:Y65B4A.6 hypothetical protein; K13025 ATP-dependent RNA he... 68.2 6e-12
cel:F33D11.10 hypothetical protein; K13025 ATP-dependent RNA h... 68.2 6e-12
bbo:BBOV_IV010990 23.m06251; eukaryotic initiation factor 4A-3... 68.2 7e-12
tgo:TGME49_056770 ATP-dependent helicase, putative (EC:3.4.22.... 67.0 1e-11
ath:AT3G19760 eukaryotic translation initiation factor 4A, put... 67.0 1e-11
tpv:TP02_0123 RNA helicase-1; K03257 translation initiation fa... 65.9 3e-11
cel:F57B9.6 inf-1; INitiation Factor family member (inf-1); K0... 64.7 6e-11
tgo:TGME49_050770 eukaryotic translation initiation factor 4A ... 64.3 1e-10
dre:394053 eif4a3, MGC56139, ddx48, eIF4A-III, zgc:56139; euka... 63.9 1e-10
mmu:434050 Gm5576, EG434050; predicted pseudogene 5576; K13025... 63.9 1e-10
xla:399362 eif4a3, XeIF-4AIII, ddx48, eif4a3-B, eif4aiii, nmp2... 63.5 1e-10
mmu:192170 Eif4a3, 2400003O03Rik, Ddx48, MGC6664, MGC6715, eIF... 63.5 1e-10
hsa:9775 EIF4A3, DDX48, DKFZp686O16189, KIAA0111, MGC10862, NM... 63.5 1e-10
dre:100330671 eukaryotic translation initiation factor 4A-like... 63.5 1e-10
cpv:cgd1_880 eukaryotic initiation factor 4A (eIF4A) (eIF-4A) 63.5 2e-10
mmu:100504817 eukaryotic initiation factor 4A-III-like; K13025... 63.5 2e-10
mmu:668137 Gm8994, B020013A22Rik, EG668137; predicted gene 899... 63.2 2e-10
bbo:BBOV_III010250 17.m07889; eukaryotic translation initiatio... 62.8 2e-10
ath:AT1G72730 eukaryotic translation initiation factor 4A, put... 61.6 6e-10
ath:AT1G54270 EIF4A-2; ATP-dependent helicase/ translation ini... 60.1 2e-09
hsa:11218 DDX20, DKFZp434H052, DP103, GEMIN3; DEAD (Asp-Glu-Al... 58.2 6e-09
ath:AT1G51380 eukaryotic translation initiation factor 4A, put... 57.8 9e-09
dre:100007313 fk48d07; wu:fk48d07; K12614 ATP-dependent RNA he... 57.4 1e-08
tpv:TP02_0613 ATP-dependent RNA helicase; K12614 ATP-dependent... 57.0 1e-08
sce:YDL160C DHH1; Cytoplasmic DExD/H-box helicase, stimulates ... 57.0 1e-08
sce:YDR021W FAL1; Fal1p (EC:3.6.1.-); K13025 ATP-dependent RNA... 57.0 1e-08
cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA he... 57.0 2e-08
cel:C07H6.5 cgh-1; Conserved Germline Helicase family member (... 56.6 2e-08
dre:556273 ddx20, wu:fb16g08, wu:fb59a11; DEAD (Asp-Glu-Ala-As... 56.2 2e-08
> tgo:TGME49_016860 ATP-dependent RNA helicase, putative ; K12812
ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=434
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 56/69 (81%), Positives = 62/69 (89%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
+ +NYDMP+S DSYLHRVGRAGRFGTKGL +TFVASQ+DTNVLNDVQTR EVHI EMP +
Sbjct 366 IVINYDMPDSSDSYLHRVGRAGRFGTKGLAITFVASQDDTNVLNDVQTRFEVHIAEMPQS 425
Query 72 IDAFQYINQ 80
IDA QYINQ
Sbjct 426 IDASQYINQ 434
> pfa:PFB0445c UAP56, U52; DEAD box helicase, UAP56; K12812 ATP-dependent
RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=457
Score = 107 bits (268), Expect = 8e-24, Method: Composition-based stats.
Identities = 51/67 (76%), Positives = 56/67 (83%), Gaps = 0/67 (0%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+NYDMPE+ DSYLHRVGRAGRFGTKGL VTFV+SQEDT LN+VQTR EV I EMP ID
Sbjct 391 INYDMPENSDSYLHRVGRAGRFGTKGLAVTFVSSQEDTLALNEVQTRFEVAISEMPNKID 450
Query 74 AFQYINQ 80
+YINQ
Sbjct 451 CNEYINQ 457
> cpv:cgd8_3900 Sub2p like superfamily II helicase involved in
snRNP biogenesis ; K12812 ATP-dependent RNA helicase UAP56/SUB2
[EC:3.6.4.13]
Length=430
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 48/69 (69%), Positives = 56/69 (81%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
+ +NYDMPE+ DSYLHRVGRAGRFGTKGL +T V+SQ D+ VLNDVQ+R EV+I EMP
Sbjct 362 IVINYDMPENTDSYLHRVGRAGRFGTKGLAITMVSSQTDSQVLNDVQSRFEVNIAEMPNQ 421
Query 72 IDAFQYINQ 80
ID YINQ
Sbjct 422 IDTSSYINQ 430
> tpv:TP03_0373 ATP-dependent RNA helicase; K12812 ATP-dependent
RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=451
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 47/67 (70%), Positives = 56/67 (83%), Gaps = 0/67 (0%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+NYDMP+S DSYLHRVGRAGRFGTKGL +TFV+S ED++ L DVQ R EV+I E+PATID
Sbjct 385 INYDMPDSTDSYLHRVGRAGRFGTKGLAITFVSSPEDSSQLEDVQKRFEVNISEIPATID 444
Query 74 AFQYINQ 80
Y+NQ
Sbjct 445 TSLYLNQ 451
> bbo:BBOV_IV002750 21.m02887; eIF-4A-like DEAD family RNA helicase
(EC:3.6.1.3); K12812 ATP-dependent RNA helicase UAP56/SUB2
[EC:3.6.4.13]
Length=472
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 44/68 (64%), Positives = 52/68 (76%), Gaps = 0/68 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
+ +NYDMP+S DSYLHRVGRAGRFGTKGL +TFVA++ D+ L DVQ R EV IPEMP +
Sbjct 392 IVINYDMPDSTDSYLHRVGRAGRFGTKGLAITFVATEADSTALADVQKRFEVDIPEMPES 451
Query 72 IDAFQYIN 79
ID Y
Sbjct 452 IDTSLYCK 459
> xla:380425 ddx39, MGC53944; nuclear RNA helicase; K13182 ATP-dependent
RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 50/66 (75%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL +TFV+ +ED +LNDVQ R EV++ E+P ID
Sbjct 360 NYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDEEDAKILNDVQDRFEVNVGELPEEIDI 419
Query 75 FQYINQ 80
YI Q
Sbjct 420 STYIEQ 425
> mmu:53817 Ddx39b, 0610030D10Rik, AI428441, Bat-1, Bat1, Bat1a,
D17H6S81E, D17H6S81E-1, D6S81Eh, MGC19235, MGC38799; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 39B (EC:3.6.4.13); K12812
ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
++ NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D +LNDVQ R EV+I E+P
Sbjct 358 IAFNYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDENDAKILNDVQDRFEVNISELPDE 417
Query 72 IDAFQYINQ 80
ID YI Q
Sbjct 418 IDISSYIEQ 426
> xla:379310 ddx39a, MGC130793, MGC53693, bat1, bat1l, ddx39,
ddxl, dxd39; DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
Length=427
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 50/66 (75%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL +TFV+ +ED +LNDVQ R EV++ E+P ID
Sbjct 360 NYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDEEDAKILNDVQDRFEVNVGELPDEIDI 419
Query 75 FQYINQ 80
YI Q
Sbjct 420 STYIEQ 425
> hsa:7919 DDX39B, BAT1, D6S81E, UAP56; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 39B (EC:3.6.4.13); K12812 ATP-dependent RNA
helicase UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
++ NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D +LNDVQ R EV+I E+P
Sbjct 358 IAFNYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDENDAKILNDVQDRFEVNISELPDE 417
Query 72 IDAFQYINQ 80
ID YI Q
Sbjct 418 IDISSYIEQ 426
> ath:AT5G11200 DEAD/DEAH box helicase, putative
Length=486
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 43/65 (66%), Positives = 50/65 (76%), Gaps = 0/65 (0%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+NYDMP+S D+YLHRVGRAGRFGTKGL +TFVAS D+ VLN VQ R EV I E+P ID
Sbjct 420 INYDMPDSADTYLHRVGRAGRFGTKGLAITFVASASDSEVLNQVQERFEVDIKELPEQID 479
Query 74 AFQYI 78
Y+
Sbjct 480 TSTYM 484
> ath:AT5G11170 ATP binding / ATP-dependent helicase/ helicase/
nucleic acid binding
Length=427
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 43/67 (64%), Positives = 51/67 (76%), Gaps = 0/67 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
+ +NYDMP+S D+YLHRVGRAGRFGTKGL +TFVAS D+ VLN VQ R EV I E+P
Sbjct 359 IVINYDMPDSADTYLHRVGRAGRFGTKGLAITFVASASDSEVLNQVQERFEVDIKELPEQ 418
Query 72 IDAFQYI 78
ID Y+
Sbjct 419 IDTSTYM 425
> dre:325550 ddx39a, wu:fc87b12, zgc:55881; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 39a
Length=346
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL VTFV+ + D +LNDVQ R EV++ E+P ID
Sbjct 279 NYDMPEDSDTYLHRVARAGRFGTKGLAVTFVSDETDAKILNDVQDRFEVNVAELPEEIDI 338
Query 75 FQYINQ 80
YI Q
Sbjct 339 STYIEQ 344
> mmu:68278 Ddx39, 2610307C23Rik, BAT1, DDXL, Ddx39a, URH49; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 39 (EC:3.6.4.13); K13182
ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL VTFV+ + D +LNDVQ R EV++ E+P ID
Sbjct 360 NYDMPEDSDTYLHRVARAGRFGTKGLAVTFVSDENDAKILNDVQDRFEVNVAELPEEIDI 419
Query 75 FQYINQ 80
YI Q
Sbjct 420 STYIEQ 425
> dre:393917 bat1, Bat1a, MGC63773, zgc:63773; HLA-B associated
transcript 1 (EC:3.6.1.-); K12812 ATP-dependent RNA helicase
UAP56/SUB2 [EC:3.6.4.13]
Length=435
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 50/69 (72%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
++ NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D LNDVQ R EV+I E+P
Sbjct 365 IAFNYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDENDARTLNDVQDRFEVNISELPEE 424
Query 72 IDAFQYINQ 80
ID YI Q
Sbjct 425 IDISSYIEQ 433
> dre:406249 ddx39b, ddx39, wu:fc16a02, zgc:55433, zgc:85646;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39b (EC:3.6.4.13); K13182
ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 41/66 (62%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D +LNDVQ R EV++ E+P ID
Sbjct 360 NYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDETDAKILNDVQDRFEVNVAELPEEIDI 419
Query 75 FQYINQ 80
YI Q
Sbjct 420 STYIEQ 425
> hsa:10212 DDX39A, BAT1, BAT1L, DDX39, DDXL, MGC18203, MGC8417,
URH49; DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A (EC:3.6.4.13);
K13182 ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=470
Score = 94.4 bits (233), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 41/66 (62%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D +LNDVQ R EV++ E+P ID
Sbjct 403 NYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDENDAKILNDVQDRFEVNVAELPEEIDI 462
Query 75 FQYINQ 80
YI Q
Sbjct 463 STYIEQ 468
> xla:447706 ddx39b, MGC81606, bat1, uap56; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 39B; K12812 ATP-dependent RNA helicase
UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 41/69 (59%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
++ NYDMPE D+YLHRV RAGRFGTKGL +TFV+ + D +LN+VQ R EV+I E+P
Sbjct 358 IAFNYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDEGDAKILNEVQDRFEVNISELPDE 417
Query 72 IDAFQYINQ 80
ID YI Q
Sbjct 418 IDISSYIEQ 426
> cel:C26D10.2 hel-1; HELicase family member (hel-1)
Length=425
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 39/64 (60%), Positives = 45/64 (70%), Gaps = 0/64 (0%)
Query 15 NYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATIDA 74
NYDMPE DSYLHRV RAGRFGTKGL +TFV+ + D LN VQ R ++ I E+P ID
Sbjct 357 NYDMPEDSDSYLHRVARAGRFGTKGLAITFVSDENDAKTLNSVQDRFDISITELPEKIDV 416
Query 75 FQYI 78
YI
Sbjct 417 STYI 420
> sce:YDL084W SUB2; Component of the TREX complex required for
nuclear mRNA export; member of the DEAD-box RNA helicase superfamily
and is involved in early and late steps of spliceosome
assembly; homolog of the human splicing factor hUAP56 (EC:3.6.1.-);
K12812 ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=446
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 48/69 (69%), Gaps = 1/69 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L++NYD+ D YLHRVGRAGRFGTKGL ++FV+S+ED VL +Q R +V I E P
Sbjct 377 LAINYDLTNEADQYLHRVGRAGRFGTKGLAISFVSSKEDEEVLAKIQERFDVKIAEFPEE 436
Query 72 -IDAFQYIN 79
ID Y+N
Sbjct 437 GIDPSTYLN 445
> pfa:PFD1070w eukaryotic initiation factor, putative; K13025
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=390
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 32/68 (47%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Query 5 LTSKELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVH 64
L +E+ L VNYD+P SR+SY+HR+GR+GRFG KG+ + FV + +D +L D++
Sbjct 320 LDVQEVSLVVNYDLPNSRESYIHRIGRSGRFGRKGVAINFVKN-DDIKILRDIEQYYSTQ 378
Query 65 IPEMPATI 72
I EMP I
Sbjct 379 IDEMPMNI 386
> tpv:TP01_0765 eukaryotic translation initiation factor 4A; K13025
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=394
Score = 68.6 bits (166), Expect = 4e-12, Method: Composition-based stats.
Identities = 30/68 (44%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query 5 LTSKELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVH 64
L +++ L VNYD+P SR+SY+HR+GR+GR+G KG+ + FV +D +L D++
Sbjct 324 LDVQQVSLVVNYDLPNSRESYIHRIGRSGRYGRKGVAINFV-KDDDIRILRDIEQYYSTQ 382
Query 65 IPEMPATI 72
I EMP I
Sbjct 383 IDEMPMNI 390
> cel:Y65B4A.6 hypothetical protein; K13025 ATP-dependent RNA
helicase [EC:3.6.4.13]
Length=399
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GRFG KG+ + FV Q+D +L D++ I EMP
Sbjct 336 LVINYDLPNNRELYIHRIGRSGRFGRKGVAINFV-KQDDVRILRDIEQYYSTQIDEMPMN 394
Query 72 I 72
I
Sbjct 395 I 395
> cel:F33D11.10 hypothetical protein; K13025 ATP-dependent RNA
helicase [EC:3.6.4.13]
Length=399
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GRFG KG+ + FV Q+D +L D++ I EMP
Sbjct 336 LVINYDLPNNRELYIHRIGRSGRFGRKGVAINFV-KQDDVRILRDIEQYYSTQIDEMPMN 394
Query 72 I 72
I
Sbjct 395 I 395
> bbo:BBOV_IV010990 23.m06251; eukaryotic initiation factor 4A-3
(eIF4A-3); K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=395
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query 5 LTSKELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVH 64
L +++ L VNYD+P SR++Y+HR+GR+GR+G KG+ + FV +D +L D++
Sbjct 325 LDVQQVSLVVNYDLPNSRENYIHRIGRSGRYGRKGVAINFVKD-DDIRILRDIEQYYSTQ 383
Query 65 IPEMPATI 72
I EMP I
Sbjct 384 IDEMPMNI 391
> tgo:TGME49_056770 ATP-dependent helicase, putative (EC:3.4.22.44);
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=395
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query 5 LTSKELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVH 64
L +++ L +NYD+P SR+ Y+HR+GR+GRFG KG+ + FV + +D +L D++
Sbjct 325 LDVQQVSLVINYDLPNSRELYIHRIGRSGRFGRKGVAINFVKN-DDIRILRDIEQYYATQ 383
Query 65 IPEMPATI 72
I EMP +
Sbjct 384 IDEMPMNV 391
> ath:AT3G19760 eukaryotic translation initiation factor 4A, putative
/ eIF-4A, putative / DEAD box RNA helicase, putative;
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=408
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 44/65 (67%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P +R+ Y+HR+GR+GRFG KG+ + FV S +D +L D++ I E
Sbjct 341 QQVSLVINYDLPNNRELYIHRIGRSGRFGRKGVAINFVKS-DDIKILRDIEQYYSTQIDE 399
Query 68 MPATI 72
MP +
Sbjct 400 MPMNV 404
> tpv:TP02_0123 RNA helicase-1; K03257 translation initiation
factor 4A
Length=400
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P S D+Y+HR+GR+GRFG KG+ + FV Q D + + ++ I E
Sbjct 333 QQVSLVINYDLPMSPDNYIHRIGRSGRFGRKGVAINFVTHQ-DMDTMKSIENYYNTQIEE 391
Query 68 MPATI 72
MPA I
Sbjct 392 MPADI 396
> cel:F57B9.6 inf-1; INitiation Factor family member (inf-1);
K03257 translation initiation factor 4A
Length=402
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 46/65 (70%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P +R++Y+HR+GR+GRFG KG+ + FV ++ D L ++++ I E
Sbjct 335 QQVSLVINYDLPSNRENYIHRIGRSGRFGRKGVAINFV-TENDARQLKEIESYYTTQIEE 393
Query 68 MPATI 72
MP +I
Sbjct 394 MPESI 398
> tgo:TGME49_050770 eukaryotic translation initiation factor 4A
(EC:3.4.22.44)
Length=412
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 44/68 (64%), Gaps = 1/68 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P ++++Y+HR+GR+GRFG KG+ + FV S D L +++ I E
Sbjct 345 QQVSLVINYDLPATKENYIHRIGRSGRFGRKGVAINFVTSS-DVEQLKEIEKHYNTQIEE 403
Query 68 MPATIDAF 75
MP + F
Sbjct 404 MPMEVAEF 411
> dre:394053 eif4a3, MGC56139, ddx48, eIF4A-III, zgc:56139; eukaryotic
translation initiation factor 4A, isoform 3 (EC:3.6.4.13);
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=406
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 45/71 (63%), Gaps = 1/71 (1%)
Query 2 AAELTSKELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRL 61
A L ++ L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++
Sbjct 333 ARGLDVSQVSLIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYY 391
Query 62 EVHIPEMPATI 72
I EMP +
Sbjct 392 STQIDEMPMNV 402
> mmu:434050 Gm5576, EG434050; predicted pseudogene 5576; K13025
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=411
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 348 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 406
Query 72 I 72
+
Sbjct 407 V 407
> xla:399362 eif4a3, XeIF-4AIII, ddx48, eif4a3-B, eif4aiii, nmp265,
nuk34; eukaryotic translation initiation factor 4A3 (EC:3.6.4.13);
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=414
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 351 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 409
Query 72 I 72
+
Sbjct 410 V 410
> mmu:192170 Eif4a3, 2400003O03Rik, Ddx48, MGC6664, MGC6715, eIF4A-III,
mKIAA0111; eukaryotic translation initiation factor
4A3 (EC:3.6.4.13); K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=411
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 348 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 406
Query 72 I 72
+
Sbjct 407 V 407
> hsa:9775 EIF4A3, DDX48, DKFZp686O16189, KIAA0111, MGC10862,
NMP265, NUK34, eIF4AIII; eukaryotic translation initiation factor
4A3 (EC:3.6.4.13); K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=411
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 348 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 406
Query 72 I 72
+
Sbjct 407 V 407
> dre:100330671 eukaryotic translation initiation factor 4A-like;
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=406
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 343 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 401
Query 72 I 72
+
Sbjct 402 V 402
> cpv:cgd1_880 eukaryotic initiation factor 4A (eIF4A) (eIF-4A)
Length=405
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P S ++Y+HR+GR+GRFG KG+ + FV + +D L D++ I E
Sbjct 337 QQVSLVINYDLPVSPETYIHRIGRSGRFGKKGVSINFV-TDDDIVCLRDIERHYNTQIEE 395
Query 68 MPATI 72
MP I
Sbjct 396 MPMGI 400
> mmu:100504817 eukaryotic initiation factor 4A-III-like; K13025
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=278
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 215 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 273
Query 72 I 72
+
Sbjct 274 L 274
> mmu:668137 Gm8994, B020013A22Rik, EG668137; predicted gene 8994;
K13025 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=411
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +NYD+P +R+ Y+HR+GR+GR+G KG+ + FV + +D +L D++ I EMP
Sbjct 348 LIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKN-DDIRILRDIEQYYSTQIDEMPMN 406
Query 72 I 72
+
Sbjct 407 L 407
> bbo:BBOV_III010250 17.m07889; eukaryotic translation initiation
factor 4A; K03257 translation initiation factor 4A
Length=402
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+P S D+Y+HR+GR+GRFG KG+ + F+ D + +++ I E
Sbjct 335 QQVSLVINYDLPMSPDNYIHRIGRSGRFGRKGVAINFLTPM-DVECMKNIENYYNTQIEE 393
Query 68 MPATI 72
MPA I
Sbjct 394 MPAEI 398
> ath:AT1G72730 eukaryotic translation initiation factor 4A, putative
/ eIF-4A, putative; K03257 translation initiation factor
4A
Length=414
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +N+D+P ++YLHR+GR+GRFG KG+ + F+ S ED ++ D+Q V + E
Sbjct 347 QQVSLVINFDLPTQPENYLHRIGRSGRFGRKGVAINFMTS-EDERMMADIQRFYNVVVEE 405
Query 68 MPATI 72
+P+ +
Sbjct 406 LPSNV 410
> ath:AT1G54270 EIF4A-2; ATP-dependent helicase/ translation initiation
factor; K03257 translation initiation factor 4A
Length=412
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +N+D+P ++YLHR+GR+GRFG KG+ + FV + +D +L D+Q V + E
Sbjct 345 QQVSLVINFDLPTQPENYLHRIGRSGRFGRKGVAINFV-TLDDQRMLFDIQKFYNVVVEE 403
Query 68 MPATI 72
+P+ +
Sbjct 404 LPSNV 408
> hsa:11218 DDX20, DKFZp434H052, DP103, GEMIN3; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 20 (EC:3.6.4.13); K13131 ATP-dependent
RNA helicase DDX20 [EC:3.6.4.13]
Length=824
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L VN D+P ++Y+HR+GRAGRFGT GL VT+ E+ N++ + + +++ +P
Sbjct 380 LVVNLDVPLDWETYMHRIGRAGRFGTLGLTVTYCCRGEEENMMMRIAQKCNINLLPLPDP 439
Query 72 IDA 74
I +
Sbjct 440 IPS 442
> ath:AT1G51380 eukaryotic translation initiation factor 4A, putative
/ eIF-4A, putative; K13025 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=392
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 38/59 (64%), Gaps = 1/59 (1%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATI 72
+NYD+P + + Y+HR+GRAGRFG +G+ + FV S D L D++ I EMPA +
Sbjct 334 INYDIPNNPELYIHRIGRAGRFGREGVAINFVKSS-DMKDLKDIERHYGTKIREMPADL 391
> dre:100007313 fk48d07; wu:fk48d07; K12614 ATP-dependent RNA
helicase DDX6/DHH1 [EC:3.6.4.13]
Length=483
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+N+D P++ ++YLHR+GR+GR+G GL + + S++ N L ++ +L I +P++ID
Sbjct 399 INFDFPKNAETYLHRIGRSGRYGHLGLAINLITSEDRFN-LKGIEDQLMTDIKPIPSSID 457
Query 74 AFQYI 78
Y+
Sbjct 458 KSLYV 462
> tpv:TP02_0613 ATP-dependent RNA helicase; K12614 ATP-dependent
RNA helicase DDX6/DHH1 [EC:3.6.4.13]
Length=417
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
VN+D P++ +YLHR+GR+GRFG GL + V Q D L ++ L I +PA +D
Sbjct 354 VNFDFPKNSSTYLHRIGRSGRFGHLGLAINLVTEQ-DKEALFKIEEELATEIKPIPAHVD 412
Query 74 AFQY 77
Y
Sbjct 413 PSLY 416
> sce:YDL160C DHH1; Cytoplasmic DExD/H-box helicase, stimulates
mRNA decapping, coordinates distinct steps in mRNA function
and decay, interacts with both the decapping and deadenylase
complexes, may have a role in mRNA export and translation
(EC:3.6.1.-); K12614 ATP-dependent RNA helicase DDX6/DHH1 [EC:3.6.4.13]
Length=506
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+N+D P++ ++YLHR+GR+GRFG GL + + + N L ++ L I +PATID
Sbjct 356 INFDFPKTAETYLHRIGRSGRFGHLGLAINLINWNDRFN-LYKIEQELGTEIAAIPATID 414
Query 74 AFQYI 78
Y+
Sbjct 415 KSLYV 419
> sce:YDR021W FAL1; Fal1p (EC:3.6.1.-); K13025 ATP-dependent RNA
helicase [EC:3.6.4.13]
Length=399
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 43/63 (68%), Gaps = 1/63 (1%)
Query 8 KELILSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPE 67
+++ L +NYD+PE ++Y+HR+GR+GRFG KG+ + F+ ++ D L +++ + I
Sbjct 332 QQVSLVINYDLPEIIENYIHRIGRSGRFGRKGVAINFI-TKADLAKLREIEKFYSIKINP 390
Query 68 MPA 70
MPA
Sbjct 391 MPA 393
> cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA
helicase DDX41 [EC:3.6.4.13]
Length=630
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMP 69
+N+DMPE ++Y+HR+GR GR G KGL TF+ + + +VL+D++ L E+P
Sbjct 514 INFDMPEDIENYVHRIGRTGRSGRKGLATTFINKKSEMSVLSDLKQLLAEAGQELP 569
> cel:C07H6.5 cgh-1; Conserved Germline Helicase family member
(cgh-1); K12614 ATP-dependent RNA helicase DDX6/DHH1 [EC:3.6.4.13]
Length=430
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Query 14 VNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPATID 73
+N+D P + ++YLHR+GR+GRFG G+ + + + ED + L ++ L I +P T+D
Sbjct 353 INFDFPRNAETYLHRIGRSGRFGHLGVAINLI-TYEDRHTLRRIEQELRTRIEPIPKTVD 411
Query 74 AFQYI 78
Y+
Sbjct 412 PKLYV 416
> dre:556273 ddx20, wu:fb16g08, wu:fb59a11; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 20 (EC:3.6.4.13); K13131 ATP-dependent
RNA helicase DDX20 [EC:3.6.4.13]
Length=788
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 12 LSVNYDMPESRDSYLHRVGRAGRFGTKGLEVTFVASQEDTNVLNDVQTRLEVHIPEMPAT 71
L +N D+P+ ++Y+HR+GRAGRFGT G+ VT+ E+ N + + + + + +P
Sbjct 374 LVINLDVPQDWETYMHRIGRAGRFGTLGVAVTYCCHGEEENKMMAIAQKCSLDLMHLPDP 433
Query 72 I 72
I
Sbjct 434 I 434
Lambda K H
0.315 0.131 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2058492688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40