bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8423_orf4
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
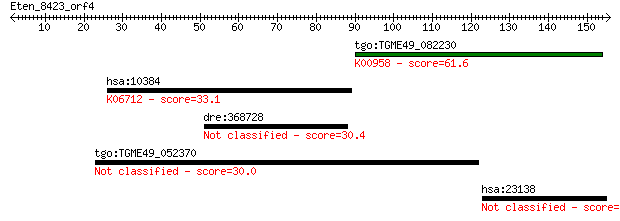
tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate k... 61.6 9e-10
hsa:10384 BTN3A3, BTF3; butyrophilin, subfamily 3, member A3; ... 33.1 0.37
dre:368728 heatr5a, CG2747, KIAA1316, MGC63665, si:busm1-142b2... 30.4 2.5
tgo:TGME49_052370 hypothetical protein 30.0 3.2
hsa:23138 N4BP3, KIAA0341; NEDD4 binding protein 3 29.6
> tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate
kinase, putative (EC:2.7.7.4 2.7.1.25); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=607
Score = 61.6 bits (148), Expect = 9e-10, Method: Composition-based stats.
Identities = 34/68 (50%), Positives = 43/68 (63%), Gaps = 5/68 (7%)
Query 90 AAAAFPRGAAAFPRGAAAQ----VGTVIAELGVESVYLPDLEEEMEKVLGTTDANHPYVK 145
A A +P A +GA + VGTVI EL V SV+ PDL+ E + VLGTTDANHPYV
Sbjct 105 ACAKYPSDCPA-AQGAVIKLRNNVGTVIVELKVSSVFEPDLQWEQQLVLGTTDANHPYVD 163
Query 146 YLRENHSE 153
Y+ N+ +
Sbjct 164 YMNTNYRD 171
> hsa:10384 BTN3A3, BTF3; butyrophilin, subfamily 3, member A3;
K06712 butyrophilin
Length=584
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 26 ASAGLGESSLLRNLLLLLQQRARLRRRSSAAAAFLRAAAAFLRGAAAFLRAAAAFLRGAA 85
S+G G S ++RN LL L++ A + S A F R+A ++ A L + L GA+
Sbjct 212 GSSGGGVSCIIRNSLLGLEKTASI----SIADPFFRSAQPWIAALAGTLPISLLLLAGAS 267
Query 86 AFL 88
FL
Sbjct 268 YFL 270
> dre:368728 heatr5a, CG2747, KIAA1316, MGC63665, si:busm1-142b24.3,
si:dz142b24.3, zgc:63665; HEAT repeat containing 5a
Length=1998
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 51 RRSSAAAAFLRAAAAFLRGAAAFLRAAAAFLRGAAAF 87
+RSS ++ FLRG A FLRA+ L+G ++
Sbjct 263 KRSSLEEVMELLSSGFLRGGAGFLRASGDMLKGTSSV 299
> tgo:TGME49_052370 hypothetical protein
Length=1990
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 44/99 (44%), Gaps = 5/99 (5%)
Query 23 RPHASAGLGESSLLRNLLLLLQQRARLRRRSSAAAAFLRAAAAFLRGAAAFLRAAAAFLR 82
RP ++G+ E + ++ + LR R +AA AA RG AAF+R ++ +
Sbjct 1696 RPKEASGMSEPHMGLSVSGGCRHETSLRWRGGDSAAHQNIYAAGFRGRAAFMRVPSSAVH 1755
Query 83 GAAAFLQAAAAFPRGAAAFPRGAAAQVGTVIAELGVESV 121
AA ++A P A G A G + L +E V
Sbjct 1756 PPAAL---SSAVPSSVLAT--GVAPAPGAALGTLSMEGV 1789
> hsa:23138 N4BP3, KIAA0341; NEDD4 binding protein 3
Length=544
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 123 LPDLEEEMEKVLGTTDANHPYVKYLRENHSEW 154
LP+ EE+++ LG D ++P+ + L E W
Sbjct 262 LPETCEELKRGLGDEDGSNPFTQVLEERQRLW 293
Lambda K H
0.323 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40