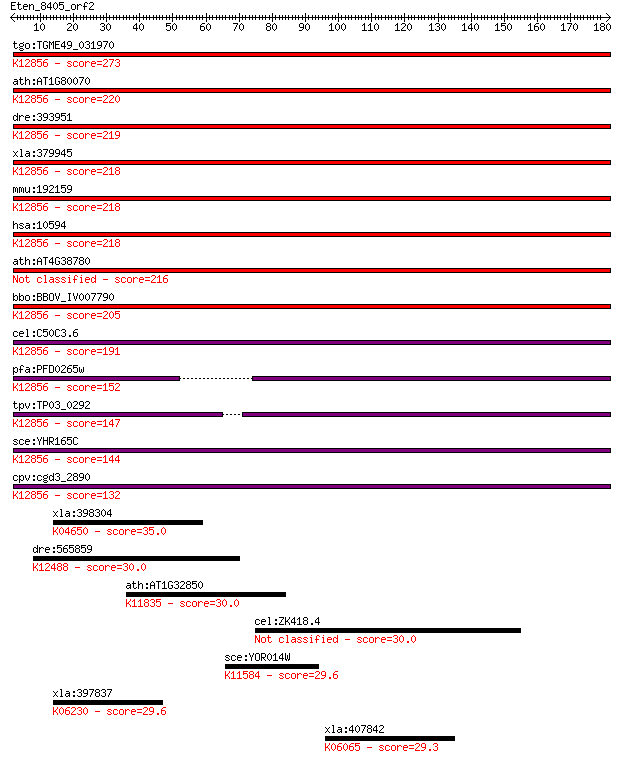
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8405_orf2
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ; K1... 273 3e-73
ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mR... 220 2e-57
dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2... 219 5e-57
xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13; P... 218 1e-56
mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfpr... 218 1e-56
hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA proce... 218 1e-56
ath:AT4G38780 splicing factor, putative 216 4e-56
bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12... 205 8e-53
cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family ... 191 1e-48
pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mR... 152 7e-37
tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing... 147 3e-35
sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component of... 144 2e-34
cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processin... 132 6e-31
xla:398304 ncor1; nuclear receptor corepressor 1; K04650 nucle... 35.0 0.13
dre:565859 asap2; ArfGAP with SH3 domain, ankyrin repeat and P... 30.0 4.0
ath:AT1G32850 UBP11; UBP11 (UBIQUITIN-SPECIFIC PROTEASE 11); c... 30.0 5.0
cel:ZK418.4 lin-37; abnormal cell LINeage family member (lin-37) 30.0 5.1
sce:YOR014W RTS1, SCS1; B-type regulatory subunit of protein p... 29.6 5.3
xla:397837 gli3, xgli3; GLI family zinc finger 3; K06230 zinc ... 29.6 6.4
xla:407842 ncor2, smrt; nuclear receptor corepressor 2; K06065... 29.3 8.3
> tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ;
K12856 pre-mRNA-processing factor 8
Length=2538
Score = 273 bits (697), Expect = 3e-73, Method: Compositional matrix adjust.
Identities = 127/180 (70%), Positives = 144/180 (80%), Gaps = 0/180 (0%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
P+LYNNRPR VA+ Y P SFVKPEDP+LPAFY+D I+NPLPAYK+ D SV
Sbjct 517 PYLYNNRPRKVAIGVYREPTCSFVKPEDPDLPAFYYDAIVNPLPAYKSGSSTTTQQDFSV 576
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
E F LPR ++ L+++PL TDTT GI LYWACRPFNLR+GR RR+ DVPLVQ+W+REH
Sbjct 577 FEDFVLPREIQPLLQDAPLSTDTTVDGIMLYWACRPFNLRSGRTRRSVDVPLVQSWYREH 636
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P NYPVKVRVSYQKLLKCWVL HLH RPPKSLKKR LFRVFK+TKFFQ TE+DWVE GL
Sbjct 637 VPTNYPVKVRVSYQKLLKCWVLNHLHQRPPKSLKKRYLFRVFKSTKFFQCTELDWVEVGL 696
> ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mRNA-processing
factor 8
Length=2382
Score = 220 bits (561), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 107/180 (59%), Positives = 130/180 (72%), Gaps = 2/180 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
PHLYNNRPR V L YHSP + ++K EDP+LPAFY+DP+I+P+ K + D
Sbjct 363 PHLYNNRPRKVKLCVYHSPMIMYIKTEDPDLPAFYYDPLIHPISNTNKEKRERKVYDDE- 421
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
+ F LP V L ++ L TDTTAAGISL +A RPFN+R+GR RRA+D+PLV WF+EH
Sbjct 422 -DDFALPEGVEPLLRDTQLYTDTTAAGISLLFAPRPFNMRSGRTRRAEDIPLVSEWFKEH 480
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P YPVKVRVSYQKLLKC+VL LH RPPK+ KK++LFR TKFFQ TE+DWVE GL
Sbjct 481 CPPAYPVKVRVSYQKLLKCYVLNELHHRPPKAQKKKHLFRSLAATKFFQSTELDWVEVGL 540
> dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2a9,
im:7141966, tdsubc_2a9, wu:fb37c02, wu:fb73e06, xx:tdsubc_2a9,
zgc:56504; pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2342
Score = 219 bits (558), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 112/180 (62%), Positives = 130/180 (72%), Gaps = 2/180 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
P+LYNN P V L YH+P V F+K EDP+LPAFYFDP+INP+ + K + L D
Sbjct 324 PYLYNNLPHHVHLTWYHTPNVVFIKTEDPDLPAFYFDPLINPISHRHSVKSQEPLPDDD- 382
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
E FELP V FL+E+PL TD TA GI+L WA RPFNLR+GR RRA DVPLV+ W+REH
Sbjct 383 -EEFELPEYVEPFLKETPLYTDNTANGIALLWAPRPFNLRSGRTRRAIDVPLVKNWYREH 441
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P PVKVRVSYQKLLK +VL L RPPK+ KKR LFR FK TKFFQ T++DWVE GL
Sbjct 442 CPAGQPVKVRVSYQKLLKYYVLNALKHRPPKAQKKRYLFRSFKATKFFQSTKLDWVEVGL 501
> xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13;
PRP8 pre-mRNA processing factor 8 homolog; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 110/180 (61%), Positives = 130/180 (72%), Gaps = 2/180 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
P+LYNN P V L YH+P V F+K EDP+LPAFYFDP+INP+ + K + L D
Sbjct 317 PYLYNNLPHHVHLTWYHTPNVVFIKTEDPDLPAFYFDPLINPISHRHSVKSQEPLPDDD- 375
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
E FELP V FL+E+PL +D TA GI+L WA RPFNLR+GR RRA D+PLV+ W+REH
Sbjct 376 -EEFELPEYVEPFLKETPLYSDNTANGIALLWAPRPFNLRSGRTRRAVDIPLVKNWYREH 434
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P PVKVRVSYQKLLK +VL L RPPK+ KKR LFR FK TKFFQ T++DWVE GL
Sbjct 435 CPAGQPVKVRVSYQKLLKYYVLNALKHRPPKAQKKRYLFRSFKATKFFQSTKLDWVEVGL 494
> mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfprp8l;
pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 110/180 (61%), Positives = 130/180 (72%), Gaps = 2/180 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
P+LYNN P V L YH+P V F+K EDP+LPAFYFDP+INP+ + K + L D
Sbjct 317 PYLYNNLPHHVHLTWYHTPNVVFIKTEDPDLPAFYFDPLINPISHRHSVKSQEPLPDDD- 375
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
E FELP V FL+++PL TD TA GI+L WA RPFNLR+GR RRA D+PLV+ W+REH
Sbjct 376 -EEFELPEFVEPFLKDTPLYTDNTANGIALLWAPRPFNLRSGRTRRALDIPLVKNWYREH 434
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P PVKVRVSYQKLLK +VL L RPPK+ KKR LFR FK TKFFQ T++DWVE GL
Sbjct 435 CPAGQPVKVRVSYQKLLKYYVLNALKHRPPKAQKKRYLFRSFKATKFFQSTKLDWVEVGL 494
> hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA processing
factor 8 homolog (S. cerevisiae); K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 110/180 (61%), Positives = 130/180 (72%), Gaps = 2/180 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSV 61
P+LYNN P V L YH+P V F+K EDP+LPAFYFDP+INP+ + K + L D
Sbjct 317 PYLYNNLPHHVHLTWYHTPNVVFIKTEDPDLPAFYFDPLINPISHRHSVKSQEPLPDDD- 375
Query 62 VEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREH 121
E FELP V FL+++PL TD TA GI+L WA RPFNLR+GR RRA D+PLV+ W+REH
Sbjct 376 -EEFELPEFVEPFLKDTPLYTDNTANGIALLWAPRPFNLRSGRTRRALDIPLVKNWYREH 434
Query 122 APQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
P PVKVRVSYQKLLK +VL L RPPK+ KKR LFR FK TKFFQ T++DWVE GL
Sbjct 435 CPAGQPVKVRVSYQKLLKYYVLNALKHRPPKAQKKRYLFRSFKATKFFQSTKLDWVEVGL 494
> ath:AT4G38780 splicing factor, putative
Length=2332
Score = 216 bits (550), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 109/187 (58%), Positives = 133/187 (71%), Gaps = 12/187 (6%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLP-AYKTNKD------FD 54
PHLYNNRPR V L YH+P V ++K EDP+LPAFY+DP+I+P+ + TNK+ +D
Sbjct 311 PHLYNNRPRKVKLCVYHTPMVMYIKTEDPDLPAFYYDPLIHPISNSNNTNKEQRKSNGYD 370
Query 55 ALADPSVVEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLV 114
D F LP + L SPL TDTTA GISL +A RPFN+R+GR RRA+D+PLV
Sbjct 371 DDGD-----DFVLPEGLEPLLNNSPLYTDTTAPGISLLFAPRPFNMRSGRTRRAEDIPLV 425
Query 115 QTWFREHAPQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEI 174
WF+EH P YPVKVRVSYQKLLKC++L LH RPPK+ KK++LFR TKFFQ TE+
Sbjct 426 AEWFKEHCPPAYPVKVRVSYQKLLKCYLLNELHHRPPKAQKKKHLFRSLAATKFFQSTEL 485
Query 175 DWVEAGL 181
DWVE GL
Sbjct 486 DWVEVGL 492
> bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12856
pre-mRNA-processing factor 8
Length=2343
Score = 205 bits (521), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 108/207 (52%), Positives = 134/207 (64%), Gaps = 27/207 (13%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAY--KTNKDFDALAD- 58
P+LY RPR VAL PYHS S++K +DP+LP FY+DPIINP+PAY K + + D +D
Sbjct 311 PYLYCIRPRKVALAPYHSKLCSYIKQDDPDLPVFYYDPIINPIPAYSIKESTELDNYSDM 370
Query 59 -------PSVVEGFELP-----------------RCVRAFLEESPLCTDTTAAGISLYWA 94
S VE + + L PL T+ T+ GI+L WA
Sbjct 371 VDNIPSKGSAVETDSMDIKPLLTQQQNVSNHNPLNGLVPLLSSVPLETENTSNGIALCWA 430
Query 95 CRPFNLRAGRMRRAQDVPLVQTWFREHAPQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSL 154
PFN R+GR RRA D+P+VQ+WFREH P ++PVKVRVSYQKLLK WVL +LHS PKS+
Sbjct 431 PHPFNKRSGRCRRAIDLPIVQSWFREHVPASHPVKVRVSYQKLLKGWVLSNLHSTRPKSM 490
Query 155 KKRNLFRVFKTTKFFQVTEIDWVEAGL 181
KKR LF+VF+ TKFFQ TE+DWVEAGL
Sbjct 491 KKRKLFKVFRATKFFQTTELDWVEAGL 517
> cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family
member (prp-8); K12856 pre-mRNA-processing factor 8
Length=2329
Score = 191 bits (485), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 103/182 (56%), Positives = 126/182 (69%), Gaps = 2/182 (1%)
Query 2 PHLYNNRPRS--VALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADP 59
P +YNN S V + YH+P+V F+K EDP+LPAFY+DP+INP+ + L +
Sbjct 305 PFMYNNLISSLPVQVSWYHTPSVVFIKTEDPDLPAFYYDPLINPIVLSNLKATEENLPEG 364
Query 60 SVVEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFR 119
+ +ELP VR E+ PL TD TA G++L WA RPFNLR+GR RRA DVPLV++W+R
Sbjct 365 EEEDEWELPEDVRPIFEDVPLYTDNTANGLALLWAPRPFNLRSGRTRRAVDVPLVKSWYR 424
Query 120 EHAPQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEA 179
EH P PVKVRVSYQKLLK +VL L RPPK K+R LFR FK TKFFQ T +DWVEA
Sbjct 425 EHCPAGMPVKVRVSYQKLLKVFVLNALKHRPPKPQKRRYLFRSFKATKFFQTTTLDWVEA 484
Query 180 GL 181
GL
Sbjct 485 GL 486
> pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mRNA-processing
factor 8
Length=3136
Score = 152 bits (383), Expect = 7e-37, Method: Composition-based stats.
Identities = 70/108 (64%), Positives = 81/108 (75%), Gaps = 0/108 (0%)
Query 74 FLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREHAPQNYPVKVRVS 133
L PL T+ T GI LY A PFN + G RR D+PLVQ+WF+EH YPVKVRVS
Sbjct 932 LLHNYPLYTERTINGIQLYHAPYPFNKKCGYTRRGIDIPLVQSWFKEHISTKYPVKVRVS 991
Query 134 YQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
YQKLLKCWVL HLHS+ PKS+KK+ LFR+FK+TKFFQ TE+DWVE GL
Sbjct 992 YQKLLKCWVLNHLHSKRPKSMKKKYLFRIFKSTKFFQCTEMDWVEVGL 1039
Score = 73.6 bits (179), Expect = 3e-13, Method: Composition-based stats.
Identities = 30/50 (60%), Positives = 38/50 (76%), Gaps = 0/50 (0%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNK 51
P+LYNNRPR +A+ YHSP ++K ED +LP FYFD IINP+P+YK K
Sbjct 663 PYLYNNRPRKIAVSKYHSPMCVYIKLEDIDLPPFYFDLIINPIPSYKIRK 712
> tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing
factor 8
Length=2736
Score = 147 bits (370), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 66/111 (59%), Positives = 84/111 (75%), Gaps = 0/111 (0%)
Query 71 VRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREHAPQNYPVKV 130
++ L L + T G+SLYWA PFN R+G RRA D+P+V +W+REH P++YPVKV
Sbjct 798 IKPLLSFVELENERTGNGVSLYWAPHPFNKRSGMCRRAIDLPIVNSWYREHVPKDYPVKV 857
Query 131 RVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEAGL 181
RVSYQKLLK WV+ +LHS+ PK +KKR LF+VF+ TKFFQ TE+DWVE GL
Sbjct 858 RVSYQKLLKGWVISNLHSKRPKGMKKRRLFKVFRGTKFFQSTELDWVEVGL 908
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 45/69 (65%), Gaps = 6/69 (8%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTN------KDFDA 55
P+LYN+RPR VA+ YH+ S+++ EDP+LP F+FDPIINP+P+Y + KD+
Sbjct 535 PYLYNSRPRKVAMANYHTKLCSYIRHEDPDLPIFHFDPIINPIPSYTIDNIDYRVKDYSI 594
Query 56 LADPSVVEG 64
S V G
Sbjct 595 KGLDSAVNG 603
> sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component
of the U4/U6-U5 snRNP complex, involved in the second catalytic
step of splicing; mutations of human Prp8 cause retinitis
pigmentosa; K12856 pre-mRNA-processing factor 8
Length=2413
Score = 144 bits (363), Expect = 2e-34, Method: Composition-based stats.
Identities = 81/182 (44%), Positives = 107/182 (58%), Gaps = 2/182 (1%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVK-PEDPELPAFYFDPIINPLPAYKTNKDFDALADPS 60
PHLYN+RPRSV +P Y++P ++ E+ + PA +FDP +NP+P + N +++
Sbjct 388 PHLYNSRPRSVRIPWYNNPVSCIIQNDEEYDTPALFFDPSLNPIPHFIDNNSSLNVSNTK 447
Query 61 VVEGFELPR-CVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFR 119
F LP EE L T +SLY + PFN G+M RAQDV L + WF
Sbjct 448 ENGDFTLPEDFAPLLAEEEELILPNTKDAMSLYHSPFPFNRTKGKMVRAQDVALAKKWFL 507
Query 120 EHAPQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEA 179
+H + YPVKV+VSYQKLLK +VL LH P + K L + K TK+FQ T IDWVEA
Sbjct 508 QHPDEEYPVKVKVSYQKLLKNYVLNELHPTLPTNHNKTKLLKSLKNTKYFQQTTIDWVEA 567
Query 180 GL 181
GL
Sbjct 568 GL 569
> cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processing
factor 8
Length=2379
Score = 132 bits (332), Expect = 6e-31, Method: Composition-based stats.
Identities = 68/182 (37%), Positives = 103/182 (56%), Gaps = 5/182 (2%)
Query 2 PHLYNNRPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPL-PAYKTNKDFDALADPS 60
PH YN+ P+ V+ YH F KPE+P P F F+ +P+ P ++ + D
Sbjct 306 PHFYNSLPKFVSTSVYHYIVNIFTKPENPNSPIFEFNEYYHPISPNNSLLENIYQIVDEE 365
Query 61 VVEGFELPRCVRAFLEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFR- 119
+ + F E L T +T GI L+W+ PFN+R+ RR+ D+ L++ W++
Sbjct 366 I---HNIKIHFSPFFHEYSLETSSTTNGILLFWSPFPFNVRSNPRRRSYDISLLKEWYKY 422
Query 120 EHAPQNYPVKVRVSYQKLLKCWVLCHLHSRPPKSLKKRNLFRVFKTTKFFQVTEIDWVEA 179
+ PVK+R+S QKLLK W+L LH++ K+ K+RN ++ + TKFFQ TE+DWVE
Sbjct 423 NNISSEQPVKIRISQQKLLKNWILNSLHNKVAKNCKRRNFLKILQNTKFFQSTEMDWVEV 482
Query 180 GL 181
GL
Sbjct 483 GL 484
> xla:398304 ncor1; nuclear receptor corepressor 1; K04650 nuclear
receptor co-repressor 1
Length=2498
Score = 35.0 bits (79), Expect = 0.13, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 14 LPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALAD 58
LPP+ PA+ F +P DP A+ F ++P P Y + A+ +
Sbjct 1619 LPPHLDPALQFHRPLDPAAAAYLFQRQLSPTPGYPSQYQLYAMEN 1663
> dre:565859 asap2; ArfGAP with SH3 domain, ankyrin repeat and
PH domain 2; K12488 Arf-GAP with SH3 domain, ANK repeat and
PH domain-containing protein
Length=1113
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 15/62 (24%), Positives = 29/62 (46%), Gaps = 0/62 (0%)
Query 8 RPRSVALPPYHSPAVSFVKPEDPELPAFYFDPIINPLPAYKTNKDFDALADPSVVEGFEL 67
+P ++ +PP SP KP+ P+ P ++P P ++++D + + L
Sbjct 983 KPITIEVPPKPSPGEVAPKPQPPDTPTIIQQGELSPQPVIQSSEDTNGSPTSAQDTPVPL 1042
Query 68 PR 69
PR
Sbjct 1043 PR 1044
> ath:AT1G32850 UBP11; UBP11 (UBIQUITIN-SPECIFIC PROTEASE 11);
cysteine-type endopeptidase/ ubiquitin thiolesterase; K11835
ubiquitin carboxyl-terminal hydrolase 4/11/15 [EC:3.1.2.15]
Length=892
Score = 30.0 bits (66), Expect = 5.0, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 4/48 (8%)
Query 36 YFDPIINPLPAYKTNKDFDALADPSVVEGFELPRCVRAFLEESPLCTD 83
Y +N LP N LA ++ EG L C+ AFL E PL D
Sbjct 708 YDSSYLNDLPKVHKN----VLAKKTMQEGISLFSCLEAFLAEEPLGPD 751
> cel:ZK418.4 lin-37; abnormal cell LINeage family member (lin-37)
Length=275
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 38/86 (44%), Gaps = 8/86 (9%)
Query 75 LEESPLCTDTTAAGISLYWACRPFNLRAGRMRRAQDVPLVQTWFREHAPQNY-PVKV-RV 132
L ESP T T + I +PF + AGR ++ L + W + H Y P+K R
Sbjct 115 LMESPRKTMTRDSKIMFELRGKPFEMIAGRF--EEEYSLGRAWVKGHMNNEYEPIKAQRT 172
Query 133 SYQ-KLLKCWVLC---HLHSRPPKSL 154
Y L ++ C H RP KS+
Sbjct 173 DYAPNLAVDYLACREIHRMPRPDKSI 198
> sce:YOR014W RTS1, SCS1; B-type regulatory subunit of protein
phosphatase 2A (PP2A); homolog of the mammalian B' subunit
of PP2A; K11584 protein phosphatase 2 (formerly 2A), regulatory
subunit B'
Length=757
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 66 ELPRCVRAFLEESPLCTDTTAAGISLYW 93
+L C+ FLE+ PL T+ G+ YW
Sbjct 515 QLAYCIVQFLEKDPLLTEEVVMGLLRYW 542
> xla:397837 gli3, xgli3; GLI family zinc finger 3; K06230 zinc
finger protein GLI
Length=1569
Score = 29.6 bits (65), Expect = 6.4, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 15/33 (45%), Gaps = 0/33 (0%)
Query 14 LPPYHSPAVSFVKPEDPELPAFYFDPIINPLPA 46
+PP H P P P+LP F P NP A
Sbjct 154 IPPLHVPTALASSPTYPDLPFFRISPHRNPASA 186
> xla:407842 ncor2, smrt; nuclear receptor corepressor 2; K06065
nuclear receptor co-repressor 2
Length=2457
Score = 29.3 bits (64), Expect = 8.3, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 96 RPFNLRAGRMRRAQDVPLVQTWFREHAPQNYPVKVRVSY 134
+P +L+ + R A P++ + E A QN PVK ++S+
Sbjct 887 KPLDLKQLKQRAAAIPPIISEGYSESAAQNTPVKQQMSH 925
Lambda K H
0.324 0.139 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40