bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8372_orf1
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
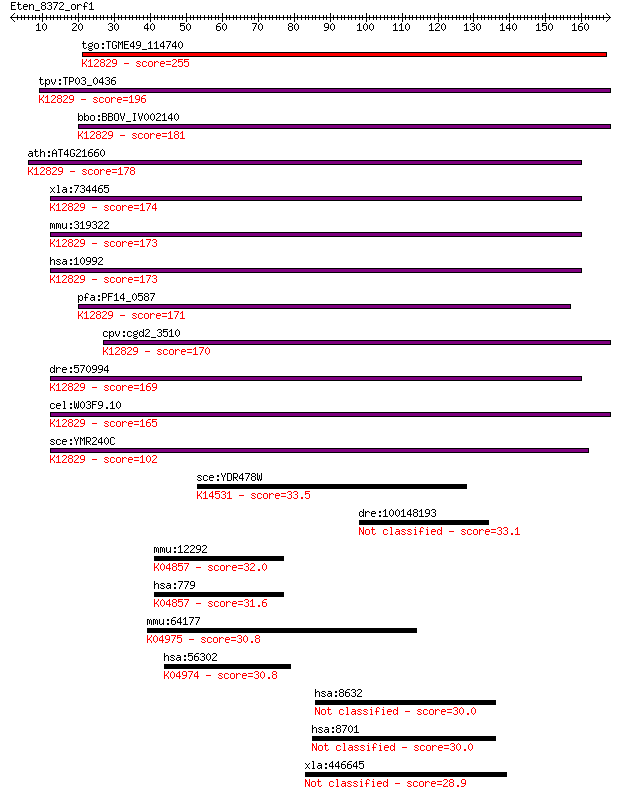
tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12... 255 5e-68
tpv:TP03_0436 hypothetical protein; K12829 splicing factor 3B ... 196 3e-50
bbo:BBOV_IV002140 21.m02783; splicing factor 3B subunit 2; K12... 181 1e-45
ath:AT4G21660 proline-rich spliceosome-associated (PSP) family... 178 7e-45
xla:734465 sf3b2, MGC115052, cus1, sap145, sf3b145, sf3b150; s... 174 1e-43
mmu:319322 Sf3b2, 145kDa, 2610311M13Rik, 2810441F20Rik, B23039... 173 2e-43
hsa:10992 SF3B2, Cus1, SAP145, SF3B145, SF3b1, SF3b150; splici... 173 2e-43
pfa:PF14_0587 splicing factor 3B subunit 2-like protein; K1282... 171 1e-42
cpv:cgd2_3510 Cus1p U2 snRNP protein ; K12829 splicing factor ... 170 2e-42
dre:570994 zgc:136773; K12829 splicing factor 3B subunit 2 169 5e-42
cel:W03F9.10 hypothetical protein; K12829 splicing factor 3B s... 165 6e-41
sce:YMR240C CUS1; Cus1p; K12829 splicing factor 3B subunit 2 102 8e-22
sce:YDR478W SNM1; Snm1p; K14531 ribonuclease MRP protein subun... 33.5 0.34
dre:100148193 dnah6; dynein, axonemal, heavy chain 6 33.1
mmu:12292 Cacna1s, AW493108, Cav1.1, Cchl1a3, fmd, mdg, sj; ca... 32.0 0.87
hsa:779 CACNA1S, CACNL1A3, CCHL1A3, Cav1.1, HOKPP, HOKPP1, MHS... 31.6 1.2
mmu:64177 Trpv6, CAT, CaT1, Cac, Ecac2, Otrpc3; transient rece... 30.8 2.1
hsa:56302 TRPV5, CAT2, ECAC1, OTRPC3; transient receptor poten... 30.8 2.1
hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489... 30.0 3.6
hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ3009... 30.0 3.7
xla:446645 ano5, MGC82557, gdd1, tmem16e; anoctamin 5 28.9
> tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12829
splicing factor 3B subunit 2
Length=743
Score = 255 bits (652), Expect = 5e-68, Method: Compositional matrix adjust.
Identities = 116/146 (79%), Positives = 126/146 (86%), Gaps = 0/146 (0%)
Query 21 RPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRGFE 80
RPS+ LKQ+T RPD +E+WDTT +DP FL +LKGLRN V VP HWSQKRRYLQWKRGFE
Sbjct 246 RPSLAELKQKTNRPDMVEIWDTTSSDPEFLVYLKGLRNTVAVPLHWSQKRRYLQWKRGFE 305
Query 81 KPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHDCFFKH 140
KPPFKLPPHIEATKISEVR+ALVEAE QK++RQRMREKVRPK NRL IDYQVLHDCFFKH
Sbjct 306 KPPFKLPPHIEATKISEVRSALVEAESQKSLRQRMREKVRPKQNRLAIDYQVLHDCFFKH 365
Query 141 AVKPPLTKMGDLYYEGKEFMQTKRNI 166
AVKP LT GDLYYEGKEF + RN
Sbjct 366 AVKPALTGFGDLYYEGKEFEKKNRNF 391
> tpv:TP03_0436 hypothetical protein; K12829 splicing factor 3B
subunit 2
Length=552
Score = 196 bits (498), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 90/159 (56%), Positives = 118/159 (74%), Gaps = 0/159 (0%)
Query 9 KRETRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQ 68
K +K +R RP++ LKQ +P+ +E+WDTT +DP FL +LKG RN V VP HWS+
Sbjct 130 KMSAKKLMRLMNRPTLYELKQSADKPEVVEIWDTTASDPKFLIWLKGQRNTVPVPSHWSE 189
Query 69 KRRYLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTI 128
K ++Q +R +KPP+KLPPHIEATKISE+R+AL E +K+++Q+ REK RPK++R+ I
Sbjct 190 KTPFMQNRRSSDKPPYKLPPHIEATKISEIRSALQIKENEKSLKQKQREKARPKSHRMDI 249
Query 129 DYQVLHDCFFKHAVKPPLTKMGDLYYEGKEFMQTKRNIK 167
DYQ LHD FFK+AVKPPLTK GD+YYEGKE RN K
Sbjct 250 DYQTLHDAFFKYAVKPPLTKYGDVYYEGKEMALRMRNCK 288
> bbo:BBOV_IV002140 21.m02783; splicing factor 3B subunit 2; K12829
splicing factor 3B subunit 2
Length=552
Score = 181 bits (458), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 82/148 (55%), Positives = 111/148 (75%), Gaps = 0/148 (0%)
Query 20 QRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRGF 79
RP++ LKQ +P+ +E WDTT ADP FL +LK RN V VP HWS K RY+Q +R +
Sbjct 146 NRPTLAQLKQMADKPEVVEFWDTTAADPRFLVWLKAQRNSVPVPSHWSDKLRYMQTRRIY 205
Query 80 EKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHDCFFK 139
+KP +KLP +IE TKI+E+R+AL++ E KT+RQ+ REKVRPK++R+ I+YQ+LHD FFK
Sbjct 206 DKPVYKLPSYIEDTKIAEIRSALIQKEANKTLRQKQREKVRPKSHRMDINYQILHDAFFK 265
Query 140 HAVKPPLTKMGDLYYEGKEFMQTKRNIK 167
+A KPP+T+ GD+YYEGKE R+ K
Sbjct 266 YATKPPMTRYGDVYYEGKEMELRMRHYK 293
> ath:AT4G21660 proline-rich spliceosome-associated (PSP) family
protein; K12829 splicing factor 3B subunit 2
Length=584
Score = 178 bits (452), Expect = 7e-45, Method: Compositional matrix adjust.
Identities = 86/154 (55%), Positives = 109/154 (70%), Gaps = 2/154 (1%)
Query 6 KKGKRETRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQH 65
+KG +KKL Q+R + LKQ +ARPD +EVWD T ADP L FLK RN V VP+H
Sbjct 149 EKGISNKKKKL--QRRMKIAELKQVSARPDVVEVWDATSADPKLLVFLKSYRNTVPVPRH 206
Query 66 WSQKRRYLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANR 125
WSQKR+YLQ KRG EK PF LP I AT I ++R A +E E K ++Q+ RE+++PK +
Sbjct 207 WSQKRKYLQGKRGIEKQPFHLPDFIAATGIEKIRQAYIEKEDGKKLKQKQRERMQPKMGK 266
Query 126 LTIDYQVLHDCFFKHAVKPPLTKMGDLYYEGKEF 159
+ IDYQVLHD FFK+ KP L+ +GDLY+EGKEF
Sbjct 267 MDIDYQVLHDAFFKYQTKPKLSALGDLYFEGKEF 300
> xla:734465 sf3b2, MGC115052, cus1, sap145, sf3b145, sf3b150;
splicing factor 3b, subunit 2, 145kDa; K12829 splicing factor
3B subunit 2
Length=764
Score = 174 bits (441), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 88/148 (59%), Positives = 105/148 (70%), Gaps = 0/148 (0%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
++KKLR+ R +V LKQ +RPD +E+ D T DP L LKG RN V VP+HW KR+
Sbjct 319 SKKKLRRMNRFTVAELKQLVSRPDVVEMHDVTAPDPKLLVHLKGTRNSVPVPRHWCFKRK 378
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQ 131
YLQ KRG EKPPF+LP I+ T I E+R AL E E+QKTM+ +MREKVRPK ++ IDYQ
Sbjct 379 YLQGKRGIEKPPFQLPDFIKRTGIQEMREALQEKEEQKTMKSKMREKVRPKMGKIDIDYQ 438
Query 132 VLHDCFFKHAVKPPLTKMGDLYYEGKEF 159
LHD FFK KP LT GDLYYEGKEF
Sbjct 439 KLHDAFFKWQSKPKLTIHGDLYYEGKEF 466
> mmu:319322 Sf3b2, 145kDa, 2610311M13Rik, 2810441F20Rik, B230398H18Rik,
SAP145, SF3b1, SF3b145, SF3b150; splicing factor
3b, subunit 2; K12829 splicing factor 3B subunit 2
Length=878
Score = 173 bits (439), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 88/148 (59%), Positives = 104/148 (70%), Gaps = 0/148 (0%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
++KKLR+ R +V LKQ ARPD +E+ D T DP L LK RN V VP+HW KR+
Sbjct 433 SKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWCFKRK 492
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQ 131
YLQ KRG EKPPF+LP I+ T I E+R AL E E+QKTM+ +MREKVRPK ++ IDYQ
Sbjct 493 YLQGKRGIEKPPFELPDFIKRTGIQEMREALQEKEEQKTMKSKMREKVRPKMGKIDIDYQ 552
Query 132 VLHDCFFKHAVKPPLTKMGDLYYEGKEF 159
LHD FFK KP LT GDLYYEGKEF
Sbjct 553 KLHDAFFKWQTKPKLTIHGDLYYEGKEF 580
> hsa:10992 SF3B2, Cus1, SAP145, SF3B145, SF3b1, SF3b150; splicing
factor 3b, subunit 2, 145kDa; K12829 splicing factor 3B
subunit 2
Length=895
Score = 173 bits (438), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 88/148 (59%), Positives = 104/148 (70%), Gaps = 0/148 (0%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
++KKLR+ R +V LKQ ARPD +E+ D T DP L LK RN V VP+HW KR+
Sbjct 450 SKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWCFKRK 509
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQ 131
YLQ KRG EKPPF+LP I+ T I E+R AL E E+QKTM+ +MREKVRPK ++ IDYQ
Sbjct 510 YLQGKRGIEKPPFELPDFIKRTGIQEMREALQEKEEQKTMKSKMREKVRPKMGKIDIDYQ 569
Query 132 VLHDCFFKHAVKPPLTKMGDLYYEGKEF 159
LHD FFK KP LT GDLYYEGKEF
Sbjct 570 KLHDAFFKWQTKPKLTIHGDLYYEGKEF 597
> pfa:PF14_0587 splicing factor 3B subunit 2-like protein; K12829
splicing factor 3B subunit 2
Length=677
Score = 171 bits (432), Expect = 1e-42, Method: Composition-based stats.
Identities = 75/137 (54%), Positives = 105/137 (76%), Gaps = 0/137 (0%)
Query 20 QRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRGF 79
+RPSV LK+ +P+ +EVWDTT +DP+F ++K L+N + VPQ W QKR+YL KRG
Sbjct 144 KRPSVMKLKEFAPKPELVEVWDTTSSDPYFYVWIKCLKNSIPVPQQWCQKRKYLHGKRGI 203
Query 80 EKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHDCFFK 139
EK P++LPP+I+ TKISE+R A+ E E+QK+++Q+MR++VRPK + + IDYQ LHD FFK
Sbjct 204 EKIPYRLPPYIQDTKISEIRQAIKEKEEQKSLKQKMRDRVRPKLHTMDIDYQTLHDAFFK 263
Query 140 HAVKPPLTKMGDLYYEG 156
+A KP L K ++YYEG
Sbjct 264 YATKPKLVKFAEVYYEG 280
> cpv:cgd2_3510 Cus1p U2 snRNP protein ; K12829 splicing factor
3B subunit 2
Length=602
Score = 170 bits (431), Expect = 2e-42, Method: Composition-based stats.
Identities = 80/141 (56%), Positives = 105/141 (74%), Gaps = 0/141 (0%)
Query 27 LKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRGFEKPPFKL 86
LK++T P+ +E+WDTT DP FL +LK V++P HW+ KRRYLQ K+G E+PP+KL
Sbjct 226 LKEKTNHPELVEIWDTTAEDPEFLVYLKSCLGSVRIPHHWNSKRRYLQGKKGLERPPYKL 285
Query 87 PPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHDCFFKHAVKPPL 146
P IE TKI+E+RA L+E E + TM+Q+ R K+RPK NR+ IDYQVLHD FF ++ KP L
Sbjct 286 PHFIEETKIAEIRALLLEEESKMTMKQKQRRKIRPKLNRMDIDYQVLHDAFFIYSTKPHL 345
Query 147 TKMGDLYYEGKEFMQTKRNIK 167
T+ GDLYYEGKEF +NI+
Sbjct 346 TQFGDLYYEGKEFEFKFKNIR 366
> dre:570994 zgc:136773; K12829 splicing factor 3B subunit 2
Length=825
Score = 169 bits (427), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 86/148 (58%), Positives = 102/148 (68%), Gaps = 0/148 (0%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
++KKLR+ R +V LKQ ARPD +E+ D T +P L LK RN V VP+HW KR+
Sbjct 375 SKKKLRRMNRLTVAELKQLVARPDVVEMHDVTAQEPKLLVHLKATRNTVPVPRHWCFKRK 434
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQ 131
YLQ KRG EKPPF+LP I T I E+R AL E E KTM+ +MREKVRPK ++ IDYQ
Sbjct 435 YLQGKRGIEKPPFELPEFIRRTGIQEMREALQEKEDAKTMKTKMREKVRPKMGKIDIDYQ 494
Query 132 VLHDCFFKHAVKPPLTKMGDLYYEGKEF 159
LHD FFK +KP LT GDLYYEGKEF
Sbjct 495 KLHDAFFKWQIKPKLTIHGDLYYEGKEF 522
> cel:W03F9.10 hypothetical protein; K12829 splicing factor 3B
subunit 2
Length=602
Score = 165 bits (418), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 78/156 (50%), Positives = 107/156 (68%), Gaps = 0/156 (0%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
+R+KLR +PS+ LK+ T R D +E D T DP+ L +K RN V VP+HW+ KR+
Sbjct 148 SRRKLRISLQPSIAKLKETTLRADVVEWADVTSRDPYLLVAMKSYRNSVPVPRHWNAKRK 207
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQ 131
YL KRGFE+PPF+LP I+ T I ++R AL+E E+ ++++ +MRE+ RPK ++ IDYQ
Sbjct 208 YLAGKRGFERPPFELPDFIKRTGIQDMREALLEKEESQSLKSKMRERARPKLGKIDIDYQ 267
Query 132 VLHDCFFKHAVKPPLTKMGDLYYEGKEFMQTKRNIK 167
LHD FFK KP +TKMG+LYYEGKE R+ K
Sbjct 268 KLHDAFFKWQTKPAMTKMGELYYEGKEMEAMMRDKK 303
> sce:YMR240C CUS1; Cus1p; K12829 splicing factor 3B subunit 2
Length=436
Score = 102 bits (253), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 56/157 (35%), Positives = 88/157 (56%), Gaps = 7/157 (4%)
Query 12 TRKKLRQQQRPSVCCLKQQTARPDRIEVWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRR 71
+ +K R+ ++PS+ LK Q P IE +D P LA +K +NV+ VP HW K+
Sbjct 122 SARKRRKTEKPSLSQLKSQVPYPQIIEWYDCDARYPGLLASIKCTKNVIPVPSHWQSKKE 181
Query 72 YLQWKRGFEKPPFKLPPHIEATKISEVRAAL----VEAEKQKTMRQRMREKVRPKANRLT 127
YL + K PF+LP I+ T I ++R+ L ++ + +K++++ R +V+PK L
Sbjct 182 YLSGRSLLGKRPFELPDIIKKTNIEQMRSTLPQSGLDGQDEKSLKEASRARVQPKMGALD 241
Query 128 IDYQVLHDCFFKHAV--KPP-LTKMGDLYYEGKEFMQ 161
+DY+ LHD FFK KP L GD+YYE + +
Sbjct 242 LDYKKLHDVFFKIGANWKPDHLLCFGDVYYENRNLFE 278
> sce:YDR478W SNM1; Snm1p; K14531 ribonuclease MRP protein subunit
SNM1
Length=198
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 3/77 (3%)
Query 53 LKGLRNVVQVPQHWSQKRRYLQWKRGFEKPPF--KLPPHIEATKISEVRAALVEAEKQKT 110
++G + +QV +K + +WK F P F + P I +T +V A+ + +K KT
Sbjct 95 MEGPKKCIQVNCLNCEKSKLFEWKSEFVVPTFGQDVSPMINSTSSGKVSYAVKKPQKSKT 154
Query 111 MRQRMREKVRPKANRLT 127
+ R K R K N LT
Sbjct 155 STGKERSKKR-KLNSLT 170
> dre:100148193 dnah6; dynein, axonemal, heavy chain 6
Length=4163
Score = 33.1 bits (74), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 98 VRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVL 133
+++ LVEAE + M R REK RP A R +I Y V+
Sbjct 3274 IKSRLVEAETTEEMINRAREKYRPVATRGSIMYFVI 3309
> mmu:12292 Cacna1s, AW493108, Cav1.1, Cchl1a3, fmd, mdg, sj;
calcium channel, voltage-dependent, L type, alpha 1S subunit;
K04857 voltage-dependent calcium channel L type alpha-1S
Length=1831
Score = 32.0 bits (71), Expect = 0.87, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 41 DTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWK 76
D G+D L ++GL ++Q +HW Q R +WK
Sbjct 389 DEGGSDTESLYEIEGLNKIIQFIRHWRQWNRVFRWK 424
> hsa:779 CACNA1S, CACNL1A3, CCHL1A3, Cav1.1, HOKPP, HOKPP1, MHS5,
TTPP1, hypoPP; calcium channel, voltage-dependent, L type,
alpha 1S subunit; K04857 voltage-dependent calcium channel
L type alpha-1S
Length=1873
Score = 31.6 bits (70), Expect = 1.2, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 41 DTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWK 76
D G+D L + GL ++Q +HW Q R +WK
Sbjct 389 DEGGSDTESLYEIAGLNKIIQFIRHWRQWNRIFRWK 424
> mmu:64177 Trpv6, CAT, CaT1, Cac, Ecac2, Otrpc3; transient receptor
potential cation channel, subfamily V, member 6; K04975
transient receptor potential cation channel subfamily V member
6
Length=727
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 36/75 (48%), Gaps = 3/75 (4%)
Query 39 VWDTTGADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRGFEKPPFKLPPHIEATKISEV 98
V + G PF LA ++G N+V QH QKR+++QW G I+++ +
Sbjct 234 VPNNQGLTPFKLAGVEG--NIVMF-QHLMQKRKHIQWTYGPLTSTLYDLTEIDSSGDDQS 290
Query 99 RAALVEAEKQKTMRQ 113
L+ K++ RQ
Sbjct 291 LLELIVTTKKREARQ 305
> hsa:56302 TRPV5, CAT2, ECAC1, OTRPC3; transient receptor potential
cation channel, subfamily V, member 5; K04974 transient
receptor potential cation channel subfamily V member 5
Length=729
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 3/35 (8%)
Query 44 GADPFFLAFLKGLRNVVQVPQHWSQKRRYLQWKRG 78
G PF LA ++G N V QH QKRR++QW G
Sbjct 240 GLTPFKLAGVEG--NTVMF-QHLMQKRRHIQWTYG 271
> hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489;
dynein, axonemal, heavy chain 17
Length=4462
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 86 LPPHIEATK--ISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHD 135
L ++E TK SE+ +VEA+ + RE RP A R ++ Y +L+D
Sbjct 3616 LVENLETTKHTASEIEEKVVEAKITEVKINEARENYRPAAERASLLYFILND 3667
> hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ30095,
FLJ37699; dynein, axonemal, heavy chain 11
Length=4523
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Query 85 KLPPHIEATK--ISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHD 135
KL +EATK ++E+ ++EA++ + RE RP A R ++ Y V++D
Sbjct 3676 KLVERLEATKTTVAEIEHKVIEAKENERKINEARECYRPVAARASLLYFVIND 3728
> xla:446645 ano5, MGC82557, gdd1, tmem16e; anoctamin 5
Length=896
Score = 28.9 bits (63), Expect = 7.7, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query 83 PFKLPPHIEATKISEVRAALVEAEKQKTMRQRMREKVRPKANRLTIDYQVLHDCFF 138
PFKLPP + + + + A +KQ+ R +EK + R I Y +L C +
Sbjct 155 PFKLPPEVMSPE-PDYFTAPFRKDKQELFRIEDKEKFFTPSTRNRIVYYILSRCHY 209
Lambda K H
0.321 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40