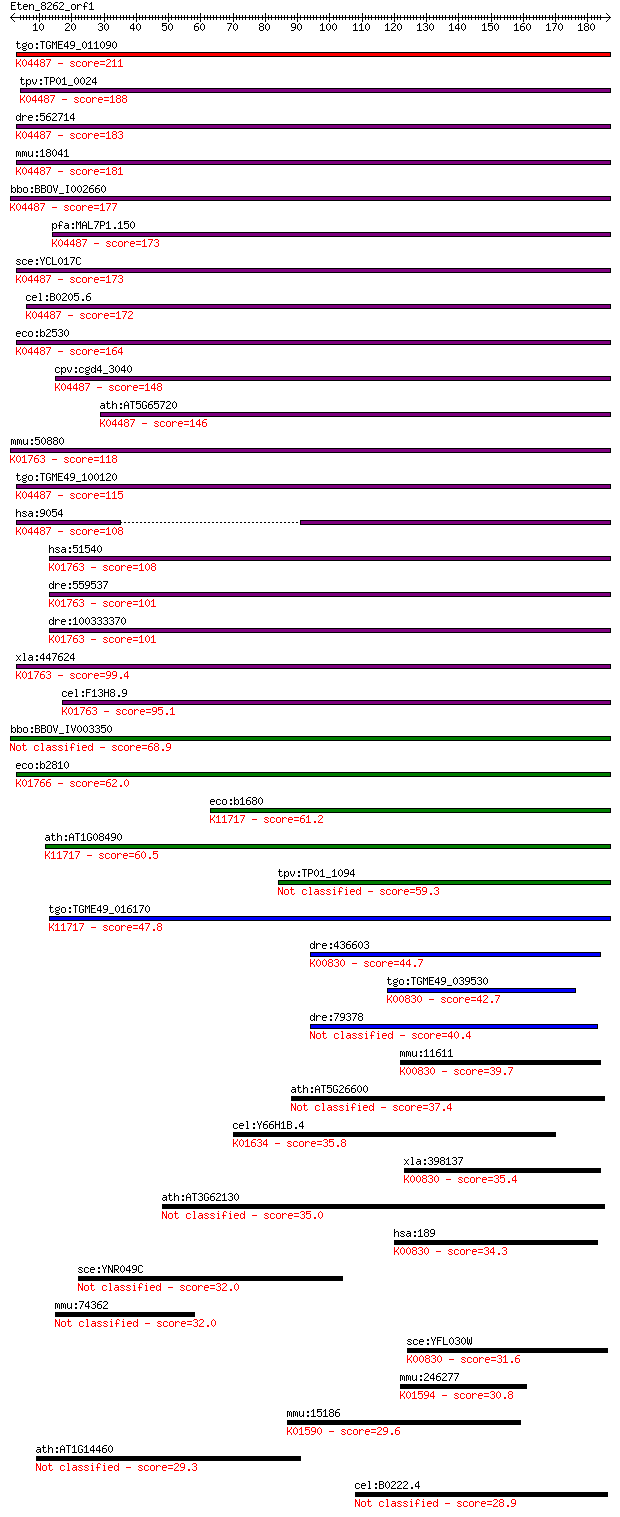
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8262_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);... 211 1e-54
tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfuras... 188 9e-48
dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation 1... 183 3e-46
mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation ge... 181 2e-45
bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);... 177 2e-44
pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-); K0... 173 2e-43
sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-... 173 3e-43
cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase ... 172 5e-43
eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine de... 164 1e-40
cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487... 148 1e-35
ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/ ... 146 3e-35
mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2; ... 118 9e-27
tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);... 115 9e-26
hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog (S... 108 9e-24
hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);... 108 1e-23
dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16... 101 1e-21
dre:100333370 selenocysteine lyase-like; K01763 selenocysteine... 101 1e-21
xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);... 99.4 7e-21
cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase ... 95.1 1e-19
bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7) 68.9 8e-12
eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desu... 62.0 1e-09
eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfur... 61.2 2e-09
ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE... 60.5 3e-09
tpv:TP01_1094 cysteine desulfurase 59.3 7e-09
tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);... 47.8 2e-05
dre:436603 agxta, agxtl, zgc:91879; alanine-glyoxylate aminotr... 44.7 2e-04
tgo:TGME49_039530 alanine--glyoxylate aminotransferase, putati... 42.7 6e-04
dre:79378 agxtb, agxt, wu:fb57d01, zgc:65930; alanine-glyoxyla... 40.4 0.004
mmu:11611 Agxt, Agt1, Agxt1; alanine-glyoxylate aminotransfera... 39.7 0.006
ath:AT5G26600 catalytic/ pyridoxal phosphate binding 37.4 0.029
cel:Y66H1B.4 spl-1; Sphingosine Phosphate Lyase family member ... 35.8 0.095
xla:398137 agxt, agt, agt1, agxt1, spt; alanine-glyoxylate ami... 35.4 0.10
ath:AT3G62130 epimerase-related 35.0 0.17
hsa:189 AGXT, AGT, AGT1, AGXT1, PH1, SPAT, SPT, TLH6; alanine-... 34.3 0.22
sce:YNR049C MSO1; Mso1p 32.0 1.2
mmu:74362 Spag17, 4931427F14Rik, PF6, Spag17-ps; sperm associa... 32.0 1.3
sce:YFL030W AGX1; Agx1p (EC:2.6.1.44); K00830 alanine-glyoxyla... 31.6 1.5
mmu:246277 Csad, Csd; cysteine sulfinic acid decarboxylase (EC... 30.8 2.6
mmu:15186 Hdc, 4732480P20, AW108189, Hdc-a, Hdc-c, Hdc-e, Hdc-... 29.6 6.3
ath:AT1G14460 DNA polymerase-related 29.3 7.4
cel:B0222.4 tag-38; Temporarily Assigned Gene name family memb... 28.9 9.7
> tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=487
Score = 211 bits (536), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 105/184 (57%), Positives = 130/184 (70%), Gaps = 20/184 (10%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA +LGL SR+REI+FTSGATESNNLA KG R + + H
Sbjct 117 VAHLLGLDASRAREIIFTSGATESNNLALKGATRAAARARRH------------------ 158
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
+ITTQ+EHKC LQCCR+L LE+ +S GA G +VT+LPV DGLV + AIRP+TL
Sbjct 159 --VITTQLEHKCALQCCRMLQLEFSESQGARGCDVTYLPVKTDGLVDLEELEKAIRPDTL 216
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
LVSV+ VNNEIGVVQ+L EIG++C+ + FHTDA+QG GK+P++VDEM IDLLS+S HK
Sbjct 217 LVSVMFVNNEIGVVQNLEEIGKICKRHDILFHTDAAQGAGKLPIDVDEMGIDLLSLSSHK 276
Query 183 IYGP 186
IYGP
Sbjct 277 IYGP 280
> tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=448
Score = 188 bits (478), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 95/183 (51%), Positives = 119/183 (65%), Gaps = 20/183 (10%)
Query 4 AAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKS 63
A + L S+ ++FTSGATESNNLA KG FY + + PGK +K+
Sbjct 92 ADIANLINCESKNVIFTSGATESNNLAIKGSKSFYGRLV------------ESPGKSKKN 139
Query 64 HIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLL 123
H+ITTQIEHKCVLQCCR L E G VT+L G++ V IRPET L
Sbjct 140 HVITTQIEHKCVLQCCRQLENE--------GYSVTYLKPDKYGMILPEEVRKNIRPETFL 191
Query 124 VSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKI 183
SVIHVNNEIGV+QD+ EIG+VC+E V FHTDA+Q FGK+P+++ + +DLLS+SGHKI
Sbjct 192 CSVIHVNNEIGVIQDIAEIGKVCKEHKVIFHTDAAQSFGKLPIDLKNLEVDLLSISGHKI 251
Query 184 YGP 186
YGP
Sbjct 252 YGP 254
> dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation
1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=451
Score = 183 bits (465), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 91/184 (49%), Positives = 120/184 (65%), Gaps = 31/184 (16%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA ++G P REIVFTSGATESNN++ KG+ RFY K +K
Sbjct 104 VAGLIGADP---REIVFTSGATESNNMSIKGVARFY--------------------KAKK 140
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
HIITTQIEHKCVL CR+L E G ++T+LPV ++GL+ + IRP+T
Sbjct 141 MHIITTQIEHKCVLDSCRVLETE--------GFDITYLPVKSNGLIDLKQLEDTIRPDTS 192
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
LVS++ +NNEIGV Q ++EIG +CR K VFFHTDA+Q GK+P++V + +DL+S+S HK
Sbjct 193 LVSIMAINNEIGVKQPVKEIGHLCRSKNVFFHTDAAQAVGKIPVDVTDWKVDLMSISAHK 252
Query 183 IYGP 186
IYGP
Sbjct 253 IYGP 256
> mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation
gene 1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=459
Score = 181 bits (458), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 91/184 (49%), Positives = 120/184 (65%), Gaps = 31/184 (16%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA+++G P REI+FTSGATESNN+A KG+ RFY + RK
Sbjct 112 VASLIGADP---REIIFTSGATESNNIAIKGVARFY--------------------RSRK 148
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
H++TTQ EHKCVL CR L A G VT+LPV G++ + AAI+P+T
Sbjct 149 KHLVTTQTEHKCVLDSCRSLE--------AEGFRVTYLPVQKSGIIDLKELEAAIQPDTS 200
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
LVSV+ VNNEIGV Q + EIG++C + V+FHTDA+Q GK+PL+V++M IDL+S+SGHK
Sbjct 201 LVSVMTVNNEIGVKQPIAEIGQICSSRKVYFHTDAAQAVGKIPLDVNDMKIDLMSISGHK 260
Query 183 IYGP 186
+YGP
Sbjct 261 LYGP 264
> bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=490
Score = 177 bits (449), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 89/186 (47%), Positives = 119/186 (63%), Gaps = 23/186 (12%)
Query 1 STVAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKP 60
S VA++L SR I+FTSGATESNNLA KG+ +Y Q + Q +
Sbjct 134 SQVASLLNCD---SRSIIFTSGATESNNLALKGITEYYSCLQHNDQSKI----------- 179
Query 61 RKSHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPE 120
K H+IT+QI+HKCVLQ CR L G VT+L G++ VAAAI P
Sbjct 180 -KDHLITSQIDHKCVLQTCRQLE--------NRGYTVTYLKPDKSGIIKPEDVAAAITPR 230
Query 121 TLLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSG 180
T + S+IH+NNEIG +Q++ IG++CREKGV FHTD +Q FGK+P+++ +N+DL+S+SG
Sbjct 231 TFMCSIIHLNNEIGTIQNIAAIGKICREKGVVFHTDGAQSFGKIPIDLSALNVDLMSISG 290
Query 181 HKIYGP 186
HKIYGP
Sbjct 291 HKIYGP 296
> pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=553
Score = 173 bits (439), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 85/173 (49%), Positives = 113/173 (65%), Gaps = 25/173 (14%)
Query 14 SREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEHK 73
++EI+FTSGATESNNLA G+ +Y + L +Q K+HIIT+QIEHK
Sbjct 207 NKEIIFTSGATESNNLALIGICTYYNK--------LNKQ---------KNHIITSQIEHK 249
Query 74 CVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHVNNEI 133
C+LQ CR L G EVT+L +GLV + +I+ T++ S I VNNEI
Sbjct 250 CILQTCRFLQ--------TKGFEVTYLKPDTNGLVKLDDIKNSIKDNTIMASFIFVNNEI 301
Query 134 GVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
GV+QD+ IG +C+EK + FHTDASQ GKVP++V +MNIDL+S+SGHK+YGP
Sbjct 302 GVIQDIENIGNLCKEKNILFHTDASQAAGKVPIDVQKMNIDLMSMSGHKLYGP 354
> sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-sulfur
cluster (Fe/S) biogenesis; required for the post-transcriptional
thio-modification of mitochondrial and cytoplasmic
tRNAs; essential protein located predominantly in mitochondria
(EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=497
Score = 173 bits (439), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 90/184 (48%), Positives = 117/184 (63%), Gaps = 31/184 (16%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA ++ P +EI+FTSGATESNN+ KG+ RFY K K
Sbjct 151 VAKMINADP---KEIIFTSGATESNNMVLKGVPRFY--------------------KKTK 187
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
HIITT+ EHKCVL+ R + E G EVTFL V GL+ + AIRP+T
Sbjct 188 KHIITTRTEHKCVLEAARAMMKE--------GFEVTFLNVDDQGLIDLKELEDAIRPDTC 239
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
LVSV+ VNNEIGV+Q ++EIG +CR+ ++FHTDA+Q +GK+ ++V+EMNIDLLS+S HK
Sbjct 240 LVSVMAVNNEIGVIQPIKEIGAICRKNKIYFHTDAAQAYGKIHIDVNEMNIDLLSISSHK 299
Query 183 IYGP 186
IYGP
Sbjct 300 IYGP 303
> cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=412
Score = 172 bits (437), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 89/181 (49%), Positives = 115/181 (63%), Gaps = 27/181 (14%)
Query 6 VLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHI 65
V L + R+I+FTSGATESNNLA KG+ +F +Q K+HI
Sbjct 64 VANLIKADPRDIIFTSGATESNNLAIKGVAKFRKQSG-------------------KNHI 104
Query 66 ITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVS 125
IT Q EHKCVL CR L E G +VT+LPV G+V + +I ET LVS
Sbjct 105 ITLQTEHKCVLDSCRYLENE--------GFKVTYLPVDKGGMVDMEQLTQSITAETCLVS 156
Query 126 VIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYG 185
++ VNNEIGV+Q +++IG +CR KGV+FHTDA+Q GKVP++V+EM IDL+S+S HKIYG
Sbjct 157 IMFVNNEIGVMQPIKQIGELCRSKGVYFHTDAAQATGKVPIDVNEMKIDLMSISAHKIYG 216
Query 186 P 186
P
Sbjct 217 P 217
> eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine
desulfurase (tRNA sulfurtransferase), PLP-dependent (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=404
Score = 164 bits (416), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 85/184 (46%), Positives = 117/184 (63%), Gaps = 31/184 (16%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
+A ++G P REIVFTSGATES+NLA KG FY Q++GK
Sbjct 58 IADLVGADP---REIVFTSGATESDNLAIKGAANFY-----------QKKGK-------- 95
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
HIIT++ EHK VL CR L E G EVT+L +G++ + AA+R +T+
Sbjct 96 -HIITSKTEHKAVLDTCRQLERE--------GFEVTYLAPQRNGIIDLKELEAAMRDDTI 146
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
LVS++HVNNEIGVVQD+ IG +CR +G+ +H DA+Q GK+P+++ ++ +DL+S SGHK
Sbjct 147 LVSIMHVNNEIGVVQDIAAIGEMCRARGIIYHVDATQSVGKLPIDLSQLKVDLMSFSGHK 206
Query 183 IYGP 186
IYGP
Sbjct 207 IYGP 210
> cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487
cysteine desulfurase [EC:2.8.1.7]
Length=438
Score = 148 bits (373), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 77/173 (44%), Positives = 106/173 (61%), Gaps = 26/173 (15%)
Query 15 REIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEHKC 74
+EI+FTSGATESNN +G+ Y + +K+HIITTQIEHKC
Sbjct 96 KEIIFTSGATESNNTIIRGVCDIYGDIEN-----------------KKNHIITTQIEHKC 138
Query 75 VLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRP-ETLLVSVIHVNNEI 133
VL R L L+ G VT+L V+ GL+S + +I P ET+L S++HVNNEI
Sbjct 139 VLSTLRELELK--------GFRVTYLKVNNKGLISLEELEKSIIPGETILASIMHVNNEI 190
Query 134 GVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
GV+Q + IG +C++ V FH+D +QG GK+ ++VD+ N D LS+S HK+YGP
Sbjct 191 GVIQPMNLIGEICKKYNVLFHSDVAQGLGKINIDVDKWNADFLSLSAHKVYGP 243
> ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/
transaminase; K04487 cysteine desulfurase [EC:2.8.1.7]
Length=325
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 76/158 (48%), Positives = 96/158 (60%), Gaps = 28/158 (17%)
Query 29 LATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEHKCVLQCCRLLHLEWQQ 88
+A KG++ FY K K H+ITTQ EHKCVL CR L E
Sbjct 1 MAVKGVMHFY--------------------KDTKKHVITTQTEHKCVLDSCRHLQQE--- 37
Query 89 SGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHVNNEIGVVQDLREIGRVCRE 148
G EVT+LPV DGLV + AIRP+T LVS++ VNNEIGVVQ + EIG +C+E
Sbjct 38 -----GFEVTYLPVKTDGLVDLEMLREAIRPDTGLVSIMAVNNEIGVVQPMEEIGMICKE 92
Query 149 KGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
V FHTDA+Q GK+P++V + N+ L+S+S HKIYGP
Sbjct 93 HNVPFHTDAAQAIGKIPVDVKKWNVALMSMSAHKIYGP 130
> mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2;
selenocysteine lyase (EC:4.4.1.16); K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=432
Score = 118 bits (296), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 72/197 (36%), Positives = 109/197 (55%), Gaps = 24/197 (12%)
Query 1 STVAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKP 60
+++A ++G P ++I+FTSG TESNNL +VR + +QQ K +++ Q +E +P
Sbjct 68 ASLAKMIGGKP---QDIIFTSGGTESNNLVIHSMVRCFHEQQTLKGNMVDQHSPEEGTRP 124
Query 61 RKSHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVS-ADGLVSAAAVAAAIRP 119
H IT +EH + L HL Q AEVTF+PVS +G + AA+RP
Sbjct 125 ---HFITCTVEHDSIR--LPLEHLVENQM-----AEVTFVPVSKVNGQAEVEDILAAVRP 174
Query 120 ETLLVSVIHVNNEIGVVQDLREIGRVCREKG----------VFFHTDASQGFGKVPLNVD 169
T LV+++ NNE GV+ + EI R + V HTDA+Q GK ++V+
Sbjct 175 TTCLVTIMLANNETGVIMPVSEISRRIKALNQIRAASGLPRVLVHTDAAQALGKRRVDVE 234
Query 170 EMNIDLLSVSGHKIYGP 186
++ +D L++ GHK YGP
Sbjct 235 DLGVDFLTIVGHKFYGP 251
> tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=851
Score = 115 bits (288), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 70/186 (37%), Positives = 99/186 (53%), Gaps = 26/186 (13%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA LG+PPS I FTSGAT N+ + ++ Q+ K G R
Sbjct 251 VAGCLGVPPS---TIFFTSGATGINSESLNWAIKCGATAQSRK------------GLDR- 294
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
HI+TT+IEH VL+ C+ L + G +VTF PV G V+ A++ +R ET
Sbjct 295 -HIVTTRIEHPAVLEICKFLEED-------HGFQVTFCPVDCFGFVNLEALSRLLRAETA 346
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKG--VFFHTDASQGFGKVPLNVDEMNIDLLSVSG 180
VSV H N EIG VQ + ++ + RE H D SQ GK+P+N+ ++ DL++++G
Sbjct 347 FVSVPHANAEIGAVQPIEKVAMIVREHAPHALLHVDCSQSLGKIPVNIPQLGADLVTIAG 406
Query 181 HKIYGP 186
HKIY P
Sbjct 407 HKIYAP 412
> hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog
(S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=406
Score = 108 bits (271), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 51/97 (52%), Positives = 70/97 (72%), Gaps = 1/97 (1%)
Query 91 GASGAEVTFLP-VSADGLVSAAAVAAAIRPETLLVSVIHVNNEIGVVQDLREIGRVCREK 149
GA E+ F + ++ + AAI+P+T LVSV+ VNNEIGV Q + EIGR+C +
Sbjct 115 GADPREIIFTSGATESNNIAIKELEAAIQPDTSLVSVMTVNNEIGVKQPIAEIGRICSSR 174
Query 150 GVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
V+FHTDA+Q GK+PL+V++M IDL+S+SGHKIYGP
Sbjct 175 KVYFHTDAAQAVGKIPLDVNDMKIDLMSISGHKIYGP 211
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 25/32 (78%), Gaps = 3/32 (9%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGL 34
VA+++G P REI+FTSGATESNN+A K L
Sbjct 110 VASLIGADP---REIIFTSGATESNNIAIKEL 138
> hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=453
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 66/185 (35%), Positives = 97/185 (52%), Gaps = 21/185 (11%)
Query 13 RSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEH 72
+ ++I+FTSG TESNNL +V+ + Q K H G P K K H IT+ +EH
Sbjct 97 KPQDIIFTSGGTESNNLVIHSVVKHFHANQTSKGH---TGGHHSPVKGAKPHFITSSVEH 153
Query 73 KCVLQCCRLLHLEWQQSGGASGAEVTFLPVS-ADGLVSAAAVAAAIRPETLLVSVIHVNN 131
+ L HL +Q A VTF+PVS G + AA+RP T LV+++ NN
Sbjct 154 DSIR--LPLEHLVEEQV-----AAVTFVPVSKVSGQAEVDDILAAVRPTTRLVTIMLANN 206
Query 132 EIGVVQDLREIGRVCRE----------KGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGH 181
E G+V + EI + + + HTDA+Q GK ++V+++ +D L++ GH
Sbjct 207 ETGIVMPVPEISQRIKALNQERVAAGLPPILVHTDAAQALGKQRVDVEDLGVDFLTIVGH 266
Query 182 KIYGP 186
K YGP
Sbjct 267 KFYGP 271
> dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=450
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 62/188 (32%), Positives = 97/188 (51%), Gaps = 27/188 (14%)
Query 13 RSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQG----KQEPGKPRKSHIITT 68
++ +I+FTSG TE+NNL F+ + K+ ++ +G KQ G+ H+I +
Sbjct 94 KAADIIFTSGGTEANNLV------FHTAVEHFKKSMISAEGSTSSKQTNGRGSLPHVIIS 147
Query 69 QIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVS-ADGLVSAAAVAAAIRPETLLVSVI 127
+EH + L E + A+VTF+PVS V V AAIRP T LVS++
Sbjct 148 NVEHDSIKLTAENLLKEGK-------ADVTFVPVSKVTARVEVEDVIAAIRPTTCLVSIM 200
Query 128 HVNNEIGVVQDLREI---------GRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSV 178
NNE G++ +++I R + HTDA+Q GK+ ++ E+ +D L++
Sbjct 201 LANNETGIIMPIKDICQRVNEVNKQRAASAPRILLHTDAAQAIGKIRVDAHELGVDYLTI 260
Query 179 SGHKIYGP 186
GHK YGP
Sbjct 261 VGHKFYGP 268
> dre:100333370 selenocysteine lyase-like; K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=450
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 62/188 (32%), Positives = 97/188 (51%), Gaps = 27/188 (14%)
Query 13 RSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQG----KQEPGKPRKSHIITT 68
++ +I+FTSG TE+NNL F+ + K+ ++ +G KQ G+ H+I +
Sbjct 94 KAADIIFTSGGTEANNLV------FHTAVEHFKKSMISAEGSTSSKQTNGRGSLPHVIIS 147
Query 69 QIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVS-ADGLVSAAAVAAAIRPETLLVSVI 127
+EH + L E + A+VTF+PVS V V AAIRP T LVS++
Sbjct 148 NVEHDSIKLTAENLLKEGK-------ADVTFVPVSKVTARVEVEDVIAAIRPTTCLVSIM 200
Query 128 HVNNEIGVVQDLREI---------GRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSV 178
NNE G++ +++I R + HTDA+Q GK+ ++ E+ +D L++
Sbjct 201 LANNETGIIMPIKDICQRVNEVNKQRAASAPRILLHTDAAQAIGKIRVDAHELGVDYLTI 260
Query 179 SGHKIYGP 186
GHK YGP
Sbjct 261 VGHKFYGP 268
> xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=426
Score = 99.4 bits (246), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 67/195 (34%), Positives = 104/195 (53%), Gaps = 29/195 (14%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
+A ++G P +I+FTSG TE+NN+ V F + ++ +Q + P
Sbjct 67 IAKMVGGKP---EDIIFTSGGTEANNM-----VLFSAVENFNRTSKERQNNNVDWALP-- 116
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSA-DGLVSAAAVAAAIRPET 121
HIIT+ +EH V LL L+ AE+TF+PVS G V V +A+RP T
Sbjct 117 -HIITSNVEHDSV--ALPLLQLQKTHK-----AEITFVPVSTVTGRVEVEDVISAVRPNT 168
Query 122 LLVSVIHVNNEIGVVQDLREIGR----VCREKG------VFFHTDASQGFGKVPLNVDEM 171
LVS++ NNE GV+ + E+ + + R++ + HTDA+Q GKV ++V E+
Sbjct 169 CLVSIMLANNETGVIMPVGELSQCLASLSRKRSAQGLPEILLHTDAAQALGKVEVDVQEL 228
Query 172 NIDLLSVSGHKIYGP 186
++ L++ GHK YGP
Sbjct 229 GVNYLTIVGHKFYGP 243
> cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase
[EC:4.4.1.16]
Length=328
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 61/174 (35%), Positives = 93/174 (53%), Gaps = 28/174 (16%)
Query 17 IVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEHKCVL 76
+VFTSG TESNN +G +R A K L HIITT IEH +L
Sbjct 9 VVFTSGGTESNNWVIEGTIR-----NAKKVSKLP-------------HIITTNIEHPSIL 50
Query 77 QCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHVNNEIGVV 136
+ + ++ G S V+ P++ G V++ ++ A+ +T LV+++ NN+ GV+
Sbjct 51 EPLK----RREEDGEISVTYVSINPLT--GFVTSQSILDALTSDTCLVTIMLANNDTGVL 104
Query 137 QDLREIGRVCREK----GVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
Q + EI + REK F H+D +Q GK+P+NV ++ D ++V GHK YGP
Sbjct 105 QPVSEIFQAIREKLKTNVPFLHSDVAQAAGKIPVNVRSLSADAVTVVGHKFYGP 158
> bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7)
Length=423
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 52/187 (27%), Positives = 81/187 (43%), Gaps = 30/187 (16%)
Query 1 STVAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKP 60
+ +A + P +R I+FTSGAT+S NL H+ PG
Sbjct 122 TAIAKFINAP--SARNIIFTSGATDSINLVANAW---------GYTHI-------RPGDT 163
Query 61 RKSHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRP- 119
++ EH + L W+ + + V F+ ++ DG + +
Sbjct 164 ----VLVPLSEHNSNI-------LPWKLLEQKNNSNVHFVKLNTDGTLDLDDYKRQLSTG 212
Query 120 ETLLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVS 179
+ L+S+ H +N +GVVQDL+ I E G DA Q V ++V ++N D L S
Sbjct 213 KVRLISIAHASNVLGVVQDLKSIIATAHEHGALVLVDACQTLAHVNIDVQQLNCDFLVAS 272
Query 180 GHKIYGP 186
GHK+YGP
Sbjct 273 GHKVYGP 279
> eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desulfinase
(EC:4.4.1.-); K01766 cysteine sulfinate desulfinase
[EC:4.4.1.-]
Length=401
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 49/184 (26%), Positives = 79/184 (42%), Gaps = 29/184 (15%)
Query 3 VAAVLGLPPSRSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRK 62
VA +L P ++ IV+T G TES N+ + R + +PG
Sbjct 72 VAQLLNAPDDKT--IVWTRGTTESINMVAQCYAR----------------PRLQPGD--- 110
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
II + EH L + W +GA+V LP++A L + I P +
Sbjct 111 -EIIVSVAEHHANL-------VPWLMVAQQTGAKVVKLPLNAQRLPDVDLLPELITPRSR 162
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
++++ ++N G DL G+ D +QG P +V +++ID + SGHK
Sbjct 163 ILALGQMSNVTGGCPDLARAITFAHSAGMVVMVDGAQGAVHFPADVQQLDIDFYAFSGHK 222
Query 183 IYGP 186
+YGP
Sbjct 223 LYGP 226
> eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfurase,
stimulated by SufE; selenocysteine lyase, PLP-dependent
(EC:4.4.1.16); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=406
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 60/124 (48%), Gaps = 7/124 (5%)
Query 63 SHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETL 122
+II +Q+EH + + WQ GAE+ +P++ DG + + +T
Sbjct 114 DNIIISQMEHHANI-------VPWQMLCARVGAELRVIPLNPDGTLQLETLPTLFDEKTR 166
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
L+++ HV+N +G L E+ + + G D +Q P++V ++ D SGHK
Sbjct 167 LLAITHVSNVLGTENPLAEMITLAHQHGAKVLVDGAQAVMHHPVDVQALDCDFYVFSGHK 226
Query 183 IYGP 186
+YGP
Sbjct 227 LYGP 230
> ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE
DESULFURASE); cysteine desulfurase/ selenocysteine lyase/
transaminase (EC:2.8.1.7); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=463
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 51/177 (28%), Positives = 75/177 (42%), Gaps = 31/177 (17%)
Query 12 SRSREIVFTSGATESNNLA--TKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQ 69
S SREIVFT ATE+ NL + GL KP I+T
Sbjct 132 SDSREIVFTRNATEAINLVAYSWGLSNL---------------------KPGDEVILTVA 170
Query 70 IEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHV 129
H C++ WQ +GA + F+ ++ D + + I P+T LV+V HV
Sbjct 171 EHHSCIVP--------WQIVSQKTGAVLKFVTLNEDEVPDINKLRELISPKTKLVAVHHV 222
Query 130 NNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
+N + + EI + G DA Q + ++V ++N D L S HK+ GP
Sbjct 223 SNVLASSLPIEEIVVWAHDVGAKVLVDACQSVPHMVVDVQKLNADFLVASSHKMCGP 279
> tpv:TP01_1094 cysteine desulfurase
Length=469
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 58/104 (55%), Gaps = 1/104 (0%)
Query 84 LEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPET-LLVSVIHVNNEIGVVQDLREI 142
L W G + ++ + +G + + ++ ++ L+ H +N +GV+QD++ I
Sbjct 188 LPWWVLCDRVGCSIEYVKLHQNGQFDLDHLESLLKSKSPKLLCCGHASNVLGVIQDMKTI 247
Query 143 GRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
++ + G +D++Q GK+ ++V +M++D L+ S HK+YGP
Sbjct 248 SKLAHKYGCLVLSDSAQTVGKIKIDVQDMDVDFLAGSSHKMYGP 291
> tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);
K11717 cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7
4.4.1.16]
Length=596
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/177 (25%), Positives = 77/177 (43%), Gaps = 30/177 (16%)
Query 13 RSREIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEH 72
R EIVFTSGAT+ NL G+ G+ I+ T EH
Sbjct 185 RPEEIVFTSGATDGINLVA------------------NTWGEANIGE--GDEIVLTIAEH 224
Query 73 KCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHVNNE 132
L + WQ A++ F+ ++ D +S +++ + + P T LV++ H +N
Sbjct 225 HSNL-------VPWQLLARRKKAQLKFVELNRDYTLSVSSLVSNLSPRTKLVALSHTSNV 277
Query 133 IGVVQD-LREIGRVCRE--KGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKIYGP 186
+G + + ++ ++ + DA+Q +V ++ D L SGHK+YGP
Sbjct 278 LGSFNPYVHHVTKLIKQINSNIVVLLDATQSLSHHQTDVRKLKCDFLVGSGHKMYGP 334
> dre:436603 agxta, agxtl, zgc:91879; alanine-glyoxylate aminotransferase
a; K00830 alanine-glyoxylate transaminase / serine-glyoxylate
transaminase / serine-pyruvate transaminase [EC:2.6.1.44
2.6.1.45 2.6.1.51]
Length=391
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 43/93 (46%), Gaps = 5/93 (5%)
Query 94 GAEVTFLPVSADGLVSAAAVAAAI---RPETLLVSVIHVNNEIGVVQDLREIGRVCREKG 150
GA+V + A G ++ + A+ RP ++ H + GVV + IG +CR+
Sbjct 118 GAKVNTVETMAGGYLTNEEIEKALNKYRPAVFFLT--HGESSTGVVHPIDGIGPLCRKYS 175
Query 151 VFFHTDASQGFGKVPLNVDEMNIDLLSVSGHKI 183
F D+ G P+ +DE ID+L K+
Sbjct 176 CLFLVDSVAALGGAPICMDEQGIDILYTGSQKV 208
> tgo:TGME49_039530 alanine--glyoxylate aminotransferase, putative
(EC:2.6.1.51); K00830 alanine-glyoxylate transaminase /
serine-glyoxylate transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=381
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 118 RPETLLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDL 175
RP ++HV GVVQ + IG +CRE D G VP+++D +DL
Sbjct 147 RPRVF--GMVHVETSTGVVQPMEGIGSLCREYDSLLLLDTVTSLGGVPVHIDAWKVDL 202
> dre:79378 agxtb, agxt, wu:fb57d01, zgc:65930; alanine-glyoxylate
aminotransferase b
Length=423
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 42/90 (46%), Gaps = 1/90 (1%)
Query 94 GAEVTFLPVSADGLVSAAAVAAAI-RPETLLVSVIHVNNEIGVVQDLREIGRVCREKGVF 152
GA+V L + G + A + A+ + + +L + H + G+V + IG VCR+
Sbjct 150 GAKVHTLAKAPGGHFTNAEIEQALAKHKPVLFFLTHGESSAGLVHPMDGIGDVCRKHNCL 209
Query 153 FHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
D+ G PL +D+ ID+L K
Sbjct 210 LLVDSVASLGAAPLLMDQQKIDILYTGSQK 239
> mmu:11611 Agxt, Agt1, Agxt1; alanine-glyoxylate aminotransferase
(EC:2.6.1.44 2.6.1.51); K00830 alanine-glyoxylate transaminase
/ serine-glyoxylate transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=414
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 122 LLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGH 181
+L+ ++H + GVVQ L G +C D+ G VP+ +D+ ID++ S
Sbjct 171 VLLFLVHGESSTGVVQPLDGFGELCHRYQCLLLVDSVASLGGVPIYMDQQGIDIMYSSSQ 230
Query 182 KI 183
K+
Sbjct 231 KV 232
> ath:AT5G26600 catalytic/ pyridoxal phosphate binding
Length=475
Score = 37.4 bits (85), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 53/107 (49%), Gaps = 14/107 (13%)
Query 88 QSGG-ASGAEVTFLPVSAD--------GLVSAAAVAAAIRPETLLVSVIHVNNEIGVVQD 138
+SGG + ++ F +SAD GL S A +R L + HV + VV
Sbjct 184 RSGGHVTEVQLPFPVISADEIIDRFRIGLESGKANGRRVR----LALIDHVTSMPSVVIP 239
Query 139 LREIGRVCREKGVF-FHTDASQGFGKVPLNVDEMNIDLLSVSGHKIY 184
++E+ ++CR +GV DA+ G G V +++ E+ D + + HK +
Sbjct 240 IKELVKICRREGVDQVFVDAAHGIGCVDVDMKEIGADFYTSNLHKWF 286
> cel:Y66H1B.4 spl-1; Sphingosine Phosphate Lyase family member
(spl-1); K01634 sphinganine-1-phosphate aldolase [EC:4.1.2.27]
Length=552
Score = 35.8 bits (81), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 42/100 (42%), Gaps = 1/100 (1%)
Query 70 IEHKCVLQCCRLLHLEWQQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPETLLVSVIHV 129
IEH +L C+ H + ++ G + +PV +D V + I ++
Sbjct 225 IEHPVIL-ACKTAHAAFDKAAHLCGMRLRHVPVDSDNRVDLKEMERLIDSNVCMLVGSAP 283
Query 130 NNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVD 169
N G + + EI ++ ++ G+ H DA G +P D
Sbjct 284 NFPSGTIDPIPEIAKLGKKYGIPVHVDACLGGFMIPFMND 323
> xla:398137 agxt, agt, agt1, agxt1, spt; alanine-glyoxylate aminotransferase;
K00830 alanine-glyoxylate transaminase / serine-glyoxylate
transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=415
Score = 35.4 bits (80), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 27/61 (44%), Gaps = 0/61 (0%)
Query 123 LVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVSGHK 182
L + H + GVVQ L +G +C D+ G P+ +D+ ID+L K
Sbjct 173 LFFITHGESSSGVVQPLDGLGDLCHRYNCLLLVDSVASLGGAPIYMDKQGIDILYSGSQK 232
Query 183 I 183
+
Sbjct 233 V 233
> ath:AT3G62130 epimerase-related
Length=454
Score = 35.0 bits (79), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 38/149 (25%), Positives = 64/149 (42%), Gaps = 19/149 (12%)
Query 48 LLQQQGK-QEPGKPRKSHIITTQIEHKCVLQCCRLLHLEWQQSGGASGAEVTF-LPVSAD 105
+LQ+ G+ GK +K T + C Q + + G S EV PV+++
Sbjct 118 VLQKVGRCFSEGKYKKE---DTVVMFHCAFQSVKKSIQAYVSRVGGSTVEVRLPFPVNSN 174
Query 106 ---------GLVSAAAVAAAIRPETLLVSVIHVNNEIGVVQDLREIGRVCREKGVF-FHT 155
GL A +R L + H+ + V+ +RE+ ++CRE+GV
Sbjct 175 EEIISKFREGLEKGRANGRTVR----LAIIDHITSMPCVLMPVRELVKICREEGVEQVFV 230
Query 156 DASQGFGKVPLNVDEMNIDLLSVSGHKIY 184
DA+ G V ++V E+ D + HK +
Sbjct 231 DAAHAIGSVKVDVKEIGADYYVSNLHKWF 259
> hsa:189 AGXT, AGT, AGT1, AGXT1, PH1, SPAT, SPT, TLH6; alanine-glyoxylate
aminotransferase (EC:2.6.1.44 2.6.1.51); K00830
alanine-glyoxylate transaminase / serine-glyoxylate transaminase
/ serine-pyruvate transaminase [EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=392
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 27/63 (42%), Gaps = 0/63 (0%)
Query 120 ETLLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVPLNVDEMNIDLLSVS 179
+ +L+ + H + GV+Q L G +C D+ G PL +D ID+L
Sbjct 147 KPVLLFLTHGESSTGVLQPLDGFGELCHRYKCLLLVDSVASLGGTPLYMDRQGIDILYSG 206
Query 180 GHK 182
K
Sbjct 207 SQK 209
> sce:YNR049C MSO1; Mso1p
Length=210
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 35/82 (42%), Gaps = 2/82 (2%)
Query 22 GATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHIITTQIEHKCVLQCCRL 81
G T + L KGLV+FYE Q Q G++E P + I+ TQ++H Q R
Sbjct 41 GDTVNTTLVHKGLVKFYENQHPF-QGFPGWLGEKE-DLPNERKILDTQVKHDMKKQNSRH 98
Query 82 LHLEWQQSGGASGAEVTFLPVS 103
+ AS + P S
Sbjct 99 FSPSFSNRRKASSEDPMGTPSS 120
> mmu:74362 Spag17, 4931427F14Rik, PF6, Spag17-ps; sperm associated
antigen 17
Length=2320
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 15 REIVFTSGATESNNLATKGLVRFYEQQQAHKQHLLQQQGKQEP 57
REI T G +ES L L+ Y AHK++ L+ Q +P
Sbjct 502 REIFLTEGESESKALPKGPLLLNYHDAHAHKKYALKDQKNFDP 544
> sce:YFL030W AGX1; Agx1p (EC:2.6.1.44); K00830 alanine-glyoxylate
transaminase / serine-glyoxylate transaminase / serine-pyruvate
transaminase [EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=385
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 27/64 (42%), Gaps = 2/64 (3%)
Query 124 VSVIHVNNEIGVVQDLREIGRVCREKG--VFFHTDASQGFGKVPLNVDEMNIDLLSVSGH 181
V+V HV+ V+ DL+ I + ++ FF DA G DE +D +
Sbjct 141 VTVTHVDTSTAVLSDLKAISQAIKQTSPETFFVVDAVCSIGCEEFEFDEWGVDFALTASQ 200
Query 182 KIYG 185
K G
Sbjct 201 KAIG 204
> mmu:246277 Csad, Csd; cysteine sulfinic acid decarboxylase (EC:4.1.1.29);
K01594 sulfinoalanine decarboxylase [EC:4.1.1.29]
Length=493
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 122 LLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQG 160
LVS +G L I VC+ G++FH DA+ G
Sbjct 239 FLVSATSGTTVLGAFDPLDAIADVCQRHGLWFHVDAAWG 277
> mmu:15186 Hdc, 4732480P20, AW108189, Hdc-a, Hdc-c, Hdc-e, Hdc-s;
histidine decarboxylase (EC:4.1.1.22); K01590 histidine
decarboxylase [EC:4.1.1.22]
Length=662
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 35/78 (44%), Gaps = 6/78 (7%)
Query 87 QQSGGASGAEVTFLPVSADGLVSAAAVAAAIRPET------LLVSVIHVNNEIGVVQDLR 140
+++G S ++ FLPV + + A+ AI + + V + L
Sbjct 205 EKAGLISLVKIRFLPVDDNFSLRGEALQKAIEEDKQQGLVPVFVCATLGTTGVCAFDRLS 264
Query 141 EIGRVCREKGVFFHTDAS 158
E+G +C +G++ H DA+
Sbjct 265 ELGPICASEGLWLHVDAA 282
> ath:AT1G14460 DNA polymerase-related
Length=1116
Score = 29.3 bits (64), Expect = 7.4, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 3/85 (3%)
Query 9 LPPSRSREI-VFTSGATES-NNLATKGLVRFYEQQQAHKQHLLQQQGKQEPGKPRKSHII 66
L ++R+I V TS TES N + K + ++Q+ + L + G Q KP ++ I+
Sbjct 953 LNSKQTRQIAVTTSSYTESGNEIPMKRIEAIIQEQRLETEWLQKTPGSQGRLKPERNQIL 1012
Query 67 TTQIEHKC-VLQCCRLLHLEWQQSG 90
+ + VL+ C + + QSG
Sbjct 1013 PQEDTNGVKVLKICEMGEFQENQSG 1037
> cel:B0222.4 tag-38; Temporarily Assigned Gene name family member
(tag-38)
Length=542
Score = 28.9 bits (63), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 36/86 (41%), Gaps = 9/86 (10%)
Query 108 VSAAAVAAAIRPETLLVSVIHVNNEIGVVQDLREIGRVCREKGVFFHTDASQGFGKVP-L 166
V + AAI T ++ N G V D+ IG++ E + H DA G +P L
Sbjct 256 VDLVKMKAAINKRTCMLVGSAPNFPFGTVDDIEAIGQLGLEYDIPVHVDACLGGFLLPFL 315
Query 167 NVDEMNIDL-------LSVSGHKIYG 185
DE+ D +S HK YG
Sbjct 316 EEDEIRYDFRVPGVSSISADSHK-YG 340
Lambda K H
0.318 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40