bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8250_orf1
Length=129
Score E
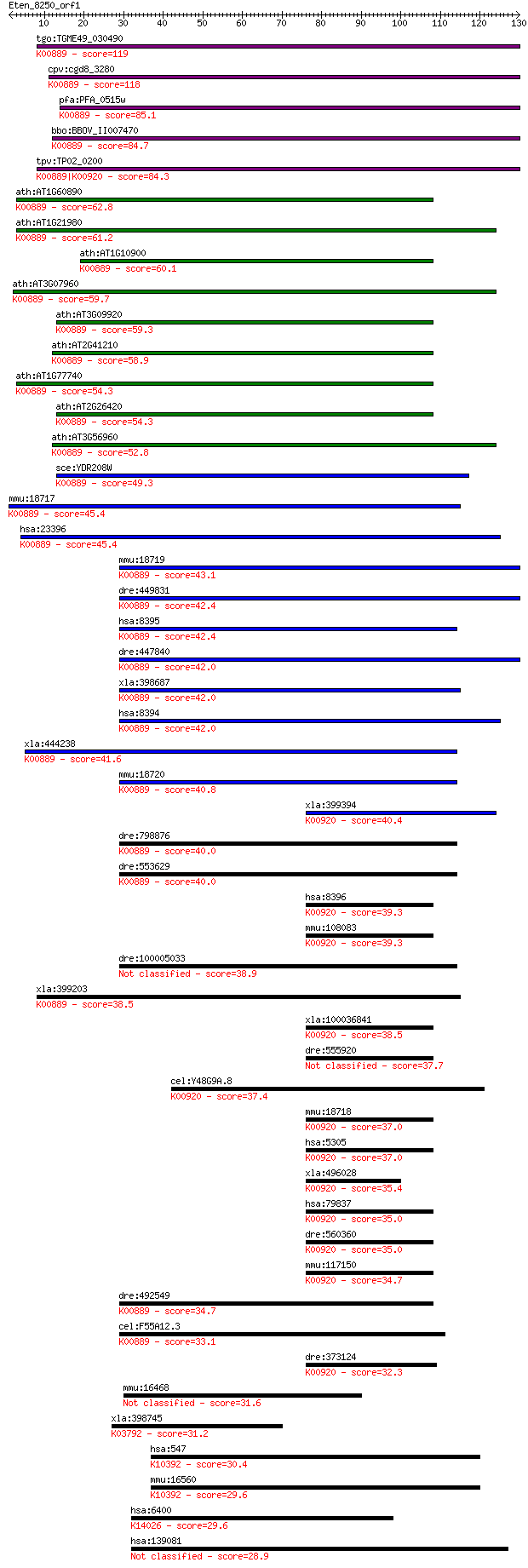
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase, p... 119 3e-27
cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K008... 118 4e-27
pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5... 85.1 5e-17
bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphat... 84.7 7e-17
tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K0088... 84.3 8e-17
ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family... 62.8 3e-10
ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KIN... 61.2 7e-10
ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family... 60.1 2e-09
ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family... 59.7 2e-09
ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 59.3 3e-09
ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHAT... 58.9 4e-09
ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, put... 54.3 9e-08
ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHA... 54.3 1e-07
ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 52.8 3e-07
sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylin... 49.3 3e-06
mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatid... 45.4 4e-05
hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma, P... 45.4 5e-05
mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7; ph... 43.1 2e-04
dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phospha... 42.4 3e-04
hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate... 42.4 3e-04
dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316... 42.0 4e-04
xla:398687 pip5k1c, MGC130766; phosphatidylinositol-4-phosphat... 42.0 5e-04
hsa:8394 PIP5K1A; phosphatidylinositol-4-phosphate 5-kinase, t... 42.0 5e-04
xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate... 41.6 7e-04
mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinosit... 40.8 0.001
xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta, ... 40.4 0.001
dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4... 40.0 0.002
dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinosito... 40.0 0.002
hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; pho... 39.3 0.003
mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosph... 39.3 0.003
dre:100005033 si:ch211-243a15.1 38.9 0.004
xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate ... 38.5 0.005
xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosp... 38.5 0.005
dre:555920 novel protein similar to vertebrate phosphatidylino... 37.7 0.009
cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920 1... 37.4 0.013
mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-p... 37.0 0.016
hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha, P... 37.0 0.016
xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate ... 35.4 0.043
hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-p... 35.0 0.057
dre:560360 similar to type II phosphatidylinositolphosphate ki... 35.0 0.059
mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate ... 34.7 0.076
dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinosit... 34.7 0.079
cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889 1... 33.1 0.21
dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phospha... 32.3 0.36
mmu:16468 Jarid2, Jmj, jumonji; jumonji, AT rich interactive d... 31.6 0.68
xla:398745 st3gal6, MGC85410, st3Gal-VI; ST3 beta-galactoside ... 31.2 0.89
hsa:547 KIF1A, ATSV, C2orf20, DKFZp686I2094, FLJ30229, HUNC-10... 30.4 1.5
mmu:16560 Kif1a, ATSV, C630002N23Rik, Gm1626, Kns1; kinesin fa... 29.6 2.2
hsa:6400 SEL1L, IBD2, PRO1063, SEL1-LIKE, SEL1L1; sel-1 suppre... 29.6 2.7
hsa:139081 MAGEC3, CT7.2, HCA2, MAGE-C3, MAGEC4, MGC119270, MG... 28.9 3.7
> tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase,
putative (EC:1.6.3.1 2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1313
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 66/123 (53%), Positives = 89/123 (72%), Gaps = 2/123 (1%)
Query 8 GQPTKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSA 67
G P KGLAV+FGH +W+ +N+M+G+RLA GR + P+R+++ YD+ KEKF +L N+
Sbjct 885 GSP-KGLAVHFGHENWDTVINIMVGIRLAAGRASSEPQRAIERYDFVMKEKFSILPNTGM 943
Query 68 LDSASALRPT-AVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCE 126
+ S S R AVRF DY+PMVFR LRE F I Y+ S+GPEQ++ N+LLG+LSSL E
Sbjct 944 VKSISDRRSLFAVRFVDYAPMVFRRLRERFHISSETYVRSVGPEQLLGNLLLGNLSSLSE 1003
Query 127 LVS 129
LVS
Sbjct 1004 LVS 1006
> cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=828
Score = 118 bits (296), Expect = 4e-27, Method: Composition-based stats.
Identities = 61/132 (46%), Positives = 82/132 (62%), Gaps = 15/132 (11%)
Query 11 TKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSS---- 66
TKG AVY GH SWN LN+M+GMRLA GRV P R + YD+ KEKF ++ +S
Sbjct 432 TKGYAVYIGHESWNTVLNMMVGMRLAIGRVYSEPNRDVADYDFIMKEKFFIIPRTSRESN 491
Query 67 ---------ALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNML 117
+ D ++P V+F DY+PMVFR +RE+ I Y+ S+GPEQ++ NM+
Sbjct 492 TKSNVLIQKSNDYRGFMKP--VQFIDYNPMVFRKIREICNISPESYVRSVGPEQLLGNMV 549
Query 118 LGSLSSLCELVS 129
LG+LSS+ EL S
Sbjct 550 LGNLSSMNELCS 561
> pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5-kinase
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1710
Score = 85.1 bits (209), Expect = 5e-17, Method: Composition-based stats.
Identities = 42/116 (36%), Positives = 72/116 (62%), Gaps = 12/116 (10%)
Query 14 LAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASA 73
LAVYFGH W++ +N+M+G+R++ S+K + N + + L +++A
Sbjct 1342 LAVYFGHERWDLVMNMMIGIRIS----------SIKKFSINDISNYFHHKDVIQLPTSNA 1391
Query 74 LRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCELVS 129
V F +Y+P++F+++R + I Y+ S+GPEQV+SNM+LG+LS+L EL+S
Sbjct 1392 QH--KVIFKNYAPIIFKNIRNFYGIKSKEYLTSVGPEQVISNMVLGNLSTLSELLS 1445
> bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphate
5-kinase (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=662
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 51/134 (38%), Positives = 81/134 (60%), Gaps = 18/134 (13%)
Query 12 KGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRL-LSNSSALDS 70
+GLAVYFGH W+ +N+M+G+ L + + + P ++KEK +S S A+D+
Sbjct 289 RGLAVYFGHERWDDVINMMVGLGLTAR--SKHEDITGDPCPDHFKEKKIFSISPSIAVDN 346
Query 71 ASALR--------PTA-------VRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSN 115
S+L PT+ V FT+++P+VF+ LR + + + YIHS+GPE +V N
Sbjct 347 MSSLNFVELESGVPTSSGDNSNRVVFTEHAPLVFKRLRALMNLSEEEYIHSVGPEHLVGN 406
Query 116 MLLGSLSSLCELVS 129
M+LG+LS+L EL+S
Sbjct 407 MVLGNLSTLSELLS 420
> tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68];
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=848
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 49/162 (30%), Positives = 74/162 (45%), Gaps = 40/162 (24%)
Query 8 GQPTKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSA 67
G +GL+V+FGH SWN LN+M+G+ ++ V L DYN K F + NS
Sbjct 445 GSSKRGLSVHFGHESWNHVLNMMIGLSISARHVYAQVNAVLSDDDYNVKLCFHINENSHG 504
Query 68 LDSASALRPTA----------------------------------------VRFTDYSPM 87
L+ + T + F +Y+PM
Sbjct 505 LNVTKFITNTGLSSSRFNSDYIINDNENGDNSLTGNNALISSNNMENVTRRIVFKEYAPM 564
Query 88 VFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCELVS 129
VFR +R + + + Y+ S+ PEQ+V NM+LG+LS++ ELVS
Sbjct 565 VFRQIRRISGLSEKDYMESVSPEQIVGNMVLGNLSTMSELVS 606
> ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=769
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 55/107 (51%), Gaps = 2/107 (1%)
Query 3 PLSITGQPTKGLAVYFGHHSWN--VALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFR 60
PL++T + T V F WN + LN+ LG+R G++ P R ++ D++ + +
Sbjct 324 PLNLTKEVTVSACVSFLGGKWNHYLMLNLQLGIRYTVGKITPVPPREVRASDFSERARIM 383
Query 61 LLSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
+ + + + DY PMVFR+LREMFK+D Y+ SI
Sbjct 384 MFFPRNGSQYTPPHKSIDFDWKDYCPMVFRNLREMFKLDAADYMMSI 430
> ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KINASE
1); 1-phosphatidylinositol-4-phosphate 5-kinase/ actin
filament binding / actin monomer binding / phosphatidylinositol
phosphate kinase; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=752
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 61/122 (50%), Gaps = 7/122 (5%)
Query 3 PLSITGQPTK-GLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRL 61
P G+ K G + GH +++ LN+ LG+R + G+ A + R LK D++ KEKF
Sbjct 334 PCWFNGEAKKPGQTISKGHKKYDLMLNLQLGIRYSVGKHA-SIVRDLKQTDFDPKEKFWT 392
Query 62 LSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSL 121
+ + R+ DY P+VFR LRE+F++D Y+ +I N L L
Sbjct 393 RFPPEGTKTTPPHQSVDFRWKDYCPLVFRRLRELFQVDPAKYMLAI-----CGNDALREL 447
Query 122 SS 123
SS
Sbjct 448 SS 449
> ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=754
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 47/89 (52%), Gaps = 0/89 (0%)
Query 19 GHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTA 78
G H++ + LN+ LG+R G++ P R ++ D+ + ++ + +
Sbjct 329 GEHNYYLMLNLQLGIRYTVGKITPVPRREVRASDFGKNARTKMFFPRDGSNFTPPHKSVD 388
Query 79 VRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
+ DY PMVFR+LR+MFK+D Y+ SI
Sbjct 389 FSWKDYCPMVFRNLRQMFKLDAAEYMMSI 417
> ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=715
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 58/122 (47%), Gaps = 5/122 (4%)
Query 2 GPLSITGQPTKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRL 61
GPL I +G + GH ++ + LN+ LG+R + GR A A LK ++ KEK
Sbjct 308 GPLRIQPAKKQGQTISKGHKNYELMLNLQLGIRHSVGRPAPATSLDLKASAFDPKEKLWT 367
Query 62 LSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSL 121
S + ++ DY P+VFR LR++F +D Y+ SI N L L
Sbjct 368 KFPSEGSKYTPPHQSCEFKWKDYCPVVFRTLRKLFSVDAADYMLSI-----CGNDALREL 422
Query 122 SS 123
SS
Sbjct 423 SS 424
> ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE); 1-phosphatidylinositol-4-phosphate 5-kinase/
ATP binding / phosphatidylinositol phosphate kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=815
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 49/95 (51%), Gaps = 0/95 (0%)
Query 13 GLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSAS 72
G V GH S+++ L++ LG+R G++ R ++ D+ + F + +
Sbjct 385 GEVVIKGHRSYDLMLSLQLGIRYTVGKITPIQRRQVRTADFGPRASFWMTFPRAGSTMTP 444
Query 73 ALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
++ DY PMVFR+LREMFKID Y+ SI
Sbjct 445 PHHSEDFKWKDYCPMVFRNLREMFKIDAADYMMSI 479
> ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHATE
5-KINASE 5); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=772
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 51/96 (53%), Gaps = 0/96 (0%)
Query 12 KGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSA 71
+G + GH ++ + LN+ LG+R + GR A A LKP ++ K+K
Sbjct 372 QGETISKGHRNYELMLNLQLGIRHSVGRQAPAASLDLKPSAFDPKDKIWRRFPREGTKYT 431
Query 72 SALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
+ T ++ DY P+VFR LR++FK+D Y+ SI
Sbjct 432 PPHQSTEFKWKDYCPLVFRSLRKLFKVDPADYMLSI 467
> ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, putative
/ PIP kinase, putative / PtdIns(4)P-5-kinase, putative
/ diphosphoinositide kinase, putative; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=754
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 53/106 (50%), Gaps = 2/106 (1%)
Query 3 PLSITGQPTK-GLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRL 61
P +G+ K G + GH +++ LN+ G+R + G+ A + R LK D++ EKF
Sbjct 336 PCCFSGEAKKPGETISKGHKKYDLMLNLQHGIRYSVGKHA-SVVRDLKQSDFDPSEKFWT 394
Query 62 LSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
+ R+ DY P+VFR LRE+F +D Y+ +I
Sbjct 395 RFPPEGSKTTPPHLSVDFRWKDYCPLVFRRLRELFTVDPADYMLAI 440
> ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHATE
5-KINASE 3); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=705
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 52/95 (54%), Gaps = 1/95 (1%)
Query 13 GLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSAS 72
G V GH ++++ LN+ LG+R + G+ A + R L+ D++ K+K S
Sbjct 319 GHTVTAGHKNYDLMLNLQLGIRYSVGKHA-SLLRELRHSDFDPKDKQWTRFPPEGSKSTP 377
Query 73 ALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
++ DY P+VFRHLR++F ID+ Y+ +I
Sbjct 378 PHLSAEFKWKDYCPIVFRHLRDLFAIDQADYMLAI 412
> ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE 4); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=779
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 54/112 (48%), Gaps = 5/112 (4%)
Query 12 KGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSA 71
+G + GH ++ + LN+ LG+R A G+ A LK ++ KEK
Sbjct 379 QGETISKGHRNYELMLNLQLGIRHAVGKQAPVVSLDLKHSAFDPKEKVWTRFPPEGTKYT 438
Query 72 SALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSS 123
+ + ++ DY P+VFR LR++FK+D Y+ SI N L LSS
Sbjct 439 PPHQSSEFKWKDYCPLVFRSLRKLFKVDPADYMLSI-----CGNDALRELSS 485
> sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=779
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 55/104 (52%), Gaps = 4/104 (3%)
Query 13 GLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSAS 72
G V GH ++ +A N++ G+R+A R + + L P D+ + +K + + L +S
Sbjct 376 GNKVSEGHVNFIIAYNMLTGIRVAVSRCS-GIMKPLTPADFRFTKKLAFDYHGNELTPSS 434
Query 73 ALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNM 116
A +F DY P VFR LR +F +D Y+ S+ + ++S +
Sbjct 435 QY---AFKFKDYCPEVFRELRALFGLDPADYLVSLTSKYILSEL 475
> mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatidylinositol-4-phosphate
5-kinase, type 1 gamma (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=635
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 54/128 (42%), Gaps = 17/128 (13%)
Query 1 DGPLSITGQPTKGLAVYFGHHSWNVALN--------------VMLGMRLAGGRVAVAPER 46
+ PL +TGQP G GH + + + LG+ G ++ PER
Sbjct 42 EAPL-VTGQPGPGHGKKLGHRGVDASGETTYKKTTSSTLKGAIQLGIGYTVGNLSSKPER 100
Query 47 SLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHS 106
+ D+ E S S L A + RF Y+P+ FR+ RE+F I Y++S
Sbjct 101 DVLMQDFYVVESIFFPSEGSNLTPAHHFQ--DFRFKTYAPVAFRYFRELFGIRPDDYLYS 158
Query 107 IGPEQVVS 114
+ E ++
Sbjct 159 LCNEPLIE 166
> hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma,
PIPKIg_v4; phosphatidylinositol-4-phosphate 5-kinase, type
I, gamma (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=640
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 56/135 (41%), Gaps = 16/135 (11%)
Query 4 LSITGQPTKGLAVYFGHHSWNVALN--------------VMLGMRLAGGRVAVAPERSLK 49
LS+T QP G GH + + + LG+ G ++ PER +
Sbjct 44 LSMTAQPGPGHGKKLGHRGVDASGETTYKKTTSSTLKGAIQLGIGYTVGHLSSKPERDVL 103
Query 50 PYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGP 109
D+ E S S L A + RF Y+P+ FR+ RE+F I Y++S+
Sbjct 104 MQDFYVVESIFFPSEGSNLTPAHHFQ--DFRFKTYAPVAFRYFRELFGIRPDDYLYSLCN 161
Query 110 EQVVSNMLLGSLSSL 124
E ++ G+ SL
Sbjct 162 EPLIELSNPGASGSL 176
> mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7;
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=539
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 46/101 (45%), Gaps = 2/101 (1%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G + PER + D+ E L S S L A RF Y+P+
Sbjct 33 IQLGIGYTVGNLTSKPERDVLMQDFYVVESVFLPSEGSNLTPAHHY--PDFRFKTYAPLA 90
Query 89 FRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCELVS 129
FR+ RE+F I Y++SI E ++ G+ SL L S
Sbjct 91 FRYFRELFGIKPDDYLYSICSEPLIELSNPGASGSLFFLTS 131
> dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=521
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 47/101 (46%), Gaps = 2/101 (1%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G + P+R + D+ E L S S L A RF +Y+P+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAHHY--PDFRFKNYAPLA 90
Query 89 FRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCELVS 129
FR+ RE+F I Y++SI E ++ G+ SS L S
Sbjct 91 FRYFRELFGIKPDDYLYSICKEPLIELSNPGASSSWFYLTS 131
> hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=540
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G + PER + D+ E L S S L A RF Y+P+
Sbjct 33 IQLGIGYTVGNLTSKPERDVLMQDFYVVESVFLPSEGSNLTPAHHY--PDFRFKTYAPLA 90
Query 89 FRHLREMFKIDKTVYIHSIGPEQVV 113
FR+ RE+F I Y++SI E ++
Sbjct 91 FRYFRELFGIKPDDYLYSICSEPLI 115
> dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=527
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 46/101 (45%), Gaps = 2/101 (1%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G + P+R + D+ E L S S L A RF Y+P+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAHHY--PDFRFKTYAPLA 90
Query 89 FRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCELVS 129
FR+ RE+F I Y++SI E ++ G+ SL L S
Sbjct 91 FRYFRELFGIKPDDYLYSICKEPLIELSNPGASGSLFYLTS 131
> xla:398687 pip5k1c, MGC130766; phosphatidylinositol-4-phosphate
5-kinase, type I, gamma (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=240
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 2/86 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G + P+R + D+ E L S S L A R RF Y+P+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAH--RYPDFRFKTYAPLA 90
Query 89 FRHLREMFKIDKTVYIHSIGPEQVVS 114
FR+ RE+F I Y++S+ E ++
Sbjct 91 FRYFRELFGIKPDDYLYSLCSEPLIE 116
> hsa:8394 PIP5K1A; phosphatidylinositol-4-phosphate 5-kinase,
type I, alpha (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=522
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 2/96 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G ++ PER + D+ E S S L A RF Y+P+
Sbjct 77 IQLGITHTVGSLSTKPERDVLMQDFYVVESIFFPSEGSNLTPAHHY--NDFRFKTYAPVA 134
Query 89 FRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSL 124
FR+ RE+F I Y++S+ E ++ G+ SL
Sbjct 135 FRYFRELFGIRPDDYLYSLCSEPLIELCSSGASGSL 170
> xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=537
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 46/109 (42%), Gaps = 2/109 (1%)
Query 5 SITGQPTKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSN 64
S T G Y S + + LG+ G + P+R + D+ E L S
Sbjct 9 SSTSDKPNGEKTYKKTTSSAIKGTIQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSE 68
Query 65 SSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVV 113
S L A RF Y+P+ FR+ RE+F I Y++S+ E ++
Sbjct 69 GSNLTPAHHY--PDFRFKTYAPLAFRYFRELFGIKPDDYLYSLCSEPLI 115
> mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinositol-4-phosphate
5-kinase, type 1 alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=546
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G ++ PER + D+ E S S L A RF Y+P+
Sbjct 74 IQLGITHTVGSLSTKPERDVLMQDFYVVESIFFPSEGSNLTPAHHY--NDFRFKTYAPVA 131
Query 89 FRHLREMFKIDKTVYIHSIGPEQVV 113
FR+ RE+F I Y++S+ E ++
Sbjct 132 FRYFRELFGIRPDDYLYSLCSEPLI 156
> xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta,
pip5k2b; phosphatidylinositol-5-phosphate 4-kinase, type II,
beta (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=417
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSS 123
P+ +F +Y PMVFR+LRE F ID Y +S+ V++ LG S
Sbjct 93 PSRFKFKEYCPMVFRNLRERFGIDDQDYQNSLTRSAPVNSENLGRFGS 140
> dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=682
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G ++ PER + D+ E S S L A RF Y+P+
Sbjct 88 IQLGIGYTVGNLSSKPERDVLMQDFYVVESIFFPSEGSNLTPAHHF--PDFRFKTYAPVA 145
Query 89 FRHLREMFKIDKTVYIHSIGPEQVV 113
FR+ RE+F I Y++S+ E ++
Sbjct 146 FRYFRELFGIRPDDYLYSLCNEPLI 170
> dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=559
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 2/85 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ + G ++ PER + D+ E S S+ S +F Y+P+
Sbjct 66 IQLGITHSVGSLSQKPERDVLMQDFEVVESIFFPSQGSS--STPGHHHGDFKFKTYAPIA 123
Query 89 FRHLREMFKIDKTVYIHSIGPEQVV 113
FR+ REMF I Y++S+ E ++
Sbjct 124 FRYFREMFGIRPDDYLYSLCNEPLI 148
> hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y PMVFR+LRE F ID Y +S+
Sbjct 90 PSRFKFKEYCPMVFRNLRERFGIDDQDYQNSV 121
> mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y PMVFR+LRE F ID Y +S+
Sbjct 90 PSRFKFKEYCPMVFRNLRERFGIDDQDYQNSV 121
> dre:100005033 si:ch211-243a15.1
Length=802
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G ++ PER + D+ E S S L A RF Y+P+
Sbjct 126 IQLGIGYTVGNLSSKPERDVLMQDFYVVESIFFPSEGSNLTPAHHY--PDFRFKTYAPVA 183
Query 89 FRHLREMFKIDKTVYIHSIGPEQVV 113
FR+ RE+F I Y++S+ E ++
Sbjct 184 FRYFRELFGIRPDDYLYSLCNEPLI 208
> xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=570
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 45/107 (42%), Gaps = 2/107 (1%)
Query 8 GQPTKGLAVYFGHHSWNVALNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSA 67
G + G Y S + + LG+ G ++ PER + D+ E S
Sbjct 46 GVDSTGETTYKKTTSSALKGAIQLGITHTVGSLSTKPERDVLMQDFYVVESIFFPGEGSN 105
Query 68 LDSASALRPTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIGPEQVVS 114
L A RF Y+P+ FR+ RE+F I Y++S+ E ++
Sbjct 106 LTPAHHY--NDFRFKTYAPVAFRYFRELFGIRPDDYLYSLCNEPLIE 150
> xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=415
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y PMVFR+LRE F ID Y +S+
Sbjct 91 PSRFKFKEYCPMVFRNLRERFGIDDQDYQNSL 122
> dre:555920 novel protein similar to vertebrate phosphatidylinositol-4-phosphate
5-kinase, type II, alpha (PIP5K2A)
Length=413
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y P+VFR+LRE F ID Y +S+
Sbjct 89 PSHFKFKEYCPLVFRNLRERFSIDDQEYQNSL 120
> cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920
1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=401
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 4/79 (5%)
Query 42 VAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRHLREMFKIDKT 101
V P L P D+ K ++ +++ D + P+ + +Y P VFR+LRE F +D
Sbjct 49 VPPPGLLMPDDFKAYSKVKIDNHNFNKD----IMPSHYKVKEYCPNVFRNLREQFGVDNF 104
Query 102 VYIHSIGPEQVVSNMLLGS 120
Y+ S+ + ++L GS
Sbjct 105 EYLRSLTSYEPEPDLLDGS 123
> mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=405
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y PMVFR+LRE F ID + +S+
Sbjct 85 PSHFKFKEYCPMVFRNLRERFGIDDQDFQNSL 116
> hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha,
PIP5KIIA, PIPK; phosphatidylinositol-5-phosphate 4-kinase,
type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=406
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y PMVFR+LRE F ID + +S+
Sbjct 85 PSHFKFKEYCPMVFRNLRERFGIDDQDFQNSL 116
> xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=419
Score = 35.4 bits (80), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKID 99
P+ +F DY P VFR+LRE F ID
Sbjct 98 PSHFKFKDYCPQVFRNLRERFGID 121
> hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 35.0 bits (79), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y P VFR+LR+ F ID Y+ S+
Sbjct 95 PSHFKFKEYCPQVFRNLRDRFGIDDQDYLVSL 126
> dre:560360 similar to type II phosphatidylinositolphosphate
kinase-alpha; K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=404
Score = 35.0 bits (79), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y P+VFR+LRE F ID + +S+
Sbjct 84 PSHFKFKEYCPLVFRNLRERFVIDDQDFQNSL 115
> mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 34.7 bits (78), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSI 107
P+ +F +Y P VFR+LR+ F ID Y+ S+
Sbjct 95 PSHFKFKEYCPQVFRNLRDRFAIDDHDYLVSL 126
> dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=584
Score = 34.7 bits (78), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMV 88
+ LG+ G ++ ER + D+ E S S L A RF Y+P+
Sbjct 64 IQLGIAHTVGSLSQKAERDVLMQDFYVVESIFFPSEGSNL--TPAHHHGDFRFKTYAPIA 121
Query 89 FRHLREMFKIDKTVYIHSI 107
FR+ RE+F I Y++S+
Sbjct 122 FRYFRELFGIRPDDYLYSL 140
> cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=611
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 42/86 (48%), Gaps = 10/86 (11%)
Query 29 VMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPT----AVRFTDY 84
+ LG+ + G +A P R + D+ EK +++ +A S + P+ RF Y
Sbjct 92 IQLGISNSIGSLASLPNRDVLLQDF---EKVDIVAFPAA---GSTITPSHSFGDFRFRTY 145
Query 85 SPMVFRHLREMFKIDKTVYIHSIGPE 110
+P+ FR+ R +F I ++ SI E
Sbjct 146 APIAFRYFRNLFHIKPADFLRSICTE 171
> dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phosphatidylinositol-4-phosphate
5-kinase, type II (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=416
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 76 PTAVRFTDYSPMVFRHLREMFKIDKTVYIHSIG 108
P+ F +Y P VFR+LRE F I+ Y S+
Sbjct 90 PSHFEFKEYCPQVFRNLRERFGIEDLDYQASLA 122
> mmu:16468 Jarid2, Jmj, jumonji; jumonji, AT rich interactive
domain 2
Length=1234
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 32/69 (46%), Gaps = 11/69 (15%)
Query 30 MLGMRLAG--------GRVAVAPERSLKPYDYNYK-EKFRLLSNSSALDSASALRPTAVR 80
MLG + G R PE S KP+D K EK S +A+D LRP+A
Sbjct 505 MLGKQAHGKTEGTPCENRSTSQPESSHKPHDPQGKPEKGSGKSGWAAMDEIPVLRPSAKE 564
Query 81 FTDYSPMVF 89
F D P+++
Sbjct 565 FHD--PLIY 571
> xla:398745 st3gal6, MGC85410, st3Gal-VI; ST3 beta-galactoside
alpha-2,3-sialyltransferase 6 (EC:2.4.99.4); K03792 beta-galactoside
alpha-2,3-sialyltransferase (sialyltransferase 10)
[EC:2.4.99.10]
Length=331
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 27 LNVMLGMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALD 69
L+ ML + + ++ P ++K + Y EKFRL+ N S +D
Sbjct 22 LHFMLHIFVQKKTISTLPTDAVKEFKTCYNEKFRLILNMSEID 64
> hsa:547 KIF1A, ATSV, C2orf20, DKFZp686I2094, FLJ30229, HUNC-104,
MGC133285, MGC133286, UNC104; kinesin family member 1A;
K10392 kinesin family member 1/13/14
Length=1690
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 38/84 (45%), Gaps = 4/84 (4%)
Query 37 GGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRHLR-EM 95
GG A +L P D NY E LS D A +R AV D + + R L+ E+
Sbjct 320 GGNSRTAMVAALSPADINYDE---TLSTLRYADRAKQIRCNAVINEDPNNKLIRELKDEV 376
Query 96 FKIDKTVYIHSIGPEQVVSNMLLG 119
++ +Y +G ++N L+G
Sbjct 377 TRLRDLLYAQGLGDITDMTNALVG 400
> mmu:16560 Kif1a, ATSV, C630002N23Rik, Gm1626, Kns1; kinesin
family member 1A; K10392 kinesin family member 1/13/14
Length=1689
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 38/84 (45%), Gaps = 4/84 (4%)
Query 37 GGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRHLR-EM 95
GG A +L P D NY E LS D A +R A+ D + + R L+ E+
Sbjct 320 GGNSRTAMVAALSPADINYDE---TLSTLRYADRAKQIRCNAIINEDPNNKLIRELKDEV 376
Query 96 FKIDKTVYIHSIGPEQVVSNMLLG 119
++ +Y +G ++N L+G
Sbjct 377 TRLRDLLYAQGLGDITDMTNALVG 400
> hsa:6400 SEL1L, IBD2, PRO1063, SEL1-LIKE, SEL1L1; sel-1 suppressor
of lin-12-like (C. elegans); K14026 SEL1 protein
Length=794
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 8/66 (12%)
Query 32 GMRLAGGRVAVAPERSLKPYDYNYKEKFRLLSNSSALDSASALRPTAVRFTDYSPMVFRH 91
GM++ G + ++S K Y Y +K ++++ AL+ S A+ F DY P +
Sbjct 190 GMKILNG----SNKKSQKREAYRYLQKAASMNHTKALERVS----YALLFGDYLPQNIQA 241
Query 92 LREMFK 97
REMF+
Sbjct 242 AREMFE 247
> hsa:139081 MAGEC3, CT7.2, HCA2, MAGE-C3, MAGEC4, MGC119270,
MGC119271; melanoma antigen family C, 3
Length=643
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 41/103 (39%), Gaps = 11/103 (10%)
Query 32 GMRLAGGRVAVAPERSLKPYDYNYKEK------FRLLSNSSA--LDSASALRPTAVRFTD 83
G RL G + PE S + Y EK F LL + L A +T
Sbjct 162 GKRLWGEKAGSLPE-SEPLFTYTLDEKVDKLVQFLLLKYQAKEPLTRAEMQMNVINTYTG 220
Query 84 YSPMVFRHLREMFKIDKTVYIHSIGPEQVVSNMLLGSLSSLCE 126
Y PM+FR RE +I + + + P+ + + +L CE
Sbjct 221 YFPMIFRKAREFIEILFGISLTEVDPDHFY--VFVNTLDLTCE 261
Lambda K H
0.320 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40