bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8205_orf1
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
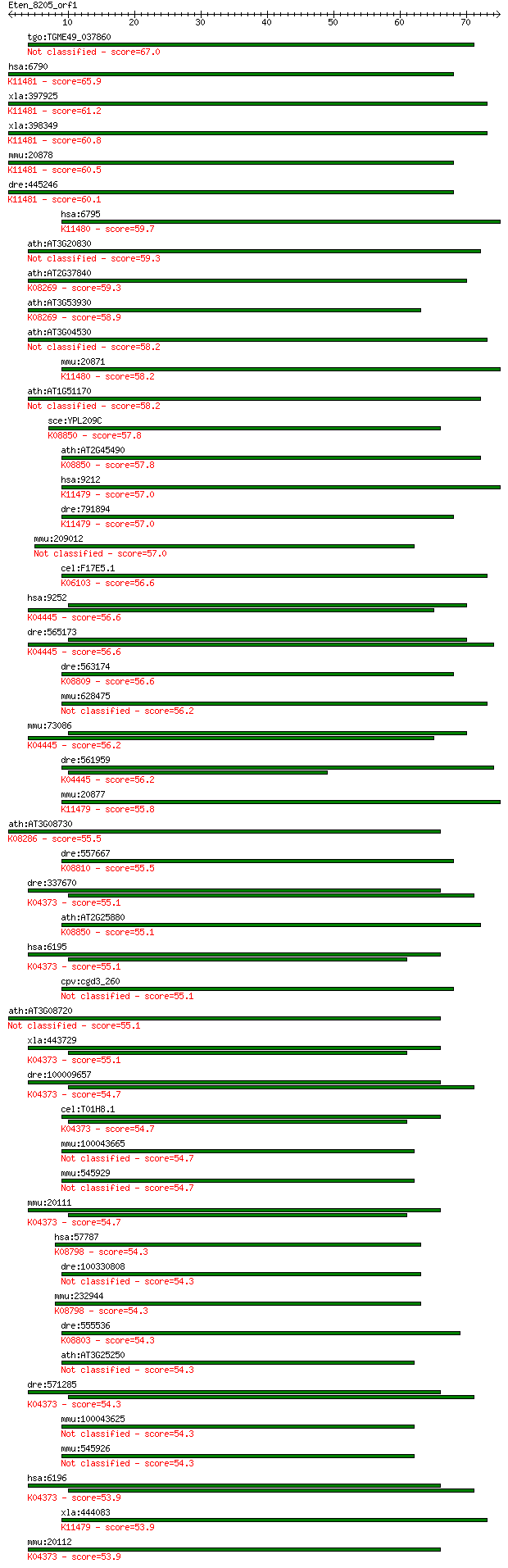
tgo:TGME49_037860 protein kinase domain-containing protein (EC... 67.0 1e-11
hsa:6790 AURKA, AIK, ARK1, AURA, AURORA2, BTAK, MGC34538, STK1... 65.9 3e-11
xla:397925 aurka, Eg2, aik, ark1, aura, aurka-a, aurora2, btak... 61.2 8e-10
xla:398349 aurka-b; p46XlEg22 (EC:2.7.11.1); K11481 aurora kin... 60.8 9e-10
mmu:20878 Aurka, AIRK1, AU019385, AW539821, Ark1, Aurora-A, Ay... 60.5 1e-09
dre:445246 aurka, zgc:100912; aurora kinase A (EC:2.7.11.1); K... 60.1 2e-09
hsa:6795 AURKC, AIE2, AIK3, ARK3, AurC, STK13, aurora-C; auror... 59.7 2e-09
ath:AT3G20830 protein kinase family protein 59.3 3e-09
ath:AT2G37840 protein kinase family protein; K08269 unc51-like... 59.3 3e-09
ath:AT3G53930 protein kinase family protein; K08269 unc51-like... 58.9 4e-09
ath:AT3G04530 PPCK2; PPCK2; kinase/ protein serine/threonine k... 58.2 7e-09
mmu:20871 Aurkc, AIE1, AIK3, IAK3, Stk13; aurora kinase C (EC:... 58.2 7e-09
ath:AT1G51170 protein kinase family protein 58.2 7e-09
sce:YPL209C IPL1, PAC15; Aurora kinase subunit of the conserve... 57.8 7e-09
ath:AT2G45490 AtAUR3; AtAUR3 (ATAURORA3); ATP binding / histon... 57.8 9e-09
hsa:9212 AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, STK12, ST... 57.0 1e-08
dre:791894 aurkb, CHUNP6896, MGC85918, fa09g06, stka, wu:fa09g... 57.0 1e-08
mmu:209012 Ulk4, 4932415A06Rik, A730098P15; unc-51-like kinase... 57.0 1e-08
cel:F17E5.1 lin-2; abnormal cell LINeage family member (lin-2)... 56.6 2e-08
hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protei... 56.6 2e-08
dre:565173 ribosomal protein S6 kinase, polypeptide 4-like; K0... 56.6 2e-08
dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific ... 56.6 2e-08
mmu:628475 Gm6882, EG628475; predicted gene 6882 56.2
mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC... 56.2 2e-08
dre:561959 si:dkey-273o13.5 (EC:2.7.11.1); K04445 mitogen-,str... 56.2 3e-08
mmu:20877 Aurkb, AIM-1, AIRK2, AL022959, Aik2, Aim1, Ark2, Aur... 55.8 3e-08
ath:AT3G08730 PK1; PK1 (PROTEIN-SERINE KINASE 1); kinase/ prot... 55.5 4e-08
dre:557667 trio; si:dkey-158b13.2 (EC:2.7.11.1); K08810 triple... 55.5 4e-08
dre:337670 rps6kal, fj94b02, rps6ka6, wu:fj94b02, zgc:66139; r... 55.1 5e-08
ath:AT2G25880 AtAUR2; AtAUR2 (AtAurora2); histone kinase(H3-S1... 55.1 5e-08
hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protei... 55.1 5e-08
cpv:cgd3_260 hypothetical protein 55.1 5e-08
ath:AT3G08720 S6K2; S6K2 (ARABIDOPSIS THALIANA SERINE/THREONIN... 55.1 5e-08
xla:443729 rps6ka6, MGC79981, rps6ka, rps6ka1; ribosomal prote... 55.1 6e-08
dre:100009657 rps6ka3b, zgc:158386; ribosomal protein S6 kinas... 54.7 6e-08
cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog famil... 54.7 6e-08
mmu:100043665 Gm10662; predicted gene 10662 54.7
mmu:545929 Gm5891, EG545929; predicted gene 5891 54.7
mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kin... 54.7 7e-08
hsa:57787 MARK4, FLJ12177, FLJ90097, KIAA1860, MARK4L, MARK4S,... 54.3 8e-08
dre:100330808 MAP/microtubule affinity-regulating kinase 3-like 54.3 9e-08
mmu:232944 Mark4, 2410090P21Rik, C79806, Markl1; MAP/microtubu... 54.3 9e-08
dre:555536 si:dkey-240h12.4; K08803 death-associated protein k... 54.3 9e-08
ath:AT3G25250 AGC2-1; AGC2-1 (OXIDATIVE SIGNAL-INDUCIBLE1); ki... 54.3 9e-08
dre:571285 ribosomal protein S6 kinase, polypeptide 2-like; K0... 54.3 9e-08
mmu:100043625 Gm4557; predicted gene 4557 54.3
mmu:545926 Gm5890, EG545926; predicted gene 5890 54.3
hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-a... 53.9 1e-07
xla:444083 aurkb-b, AurB, MGC83575, aik2, aim1, airk2, ark2, i... 53.9 1e-07
mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk... 53.9 1e-07
> tgo:TGME49_037860 protein kinase domain-containing protein (EC:2.7.11.1
2.7.10.2)
Length=2073
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A++ GTL+YAAPE++ G YD + D+WS+GV++FELL+ G LPF G T +++ I+
Sbjct 1946 ASDSLGTLAYAAPEILVGLEYDKAVDMWSIGVITFELLSQGVLPFQGKTEQQIGASILEG 2005
Query 64 EIDFEGG 70
F+ G
Sbjct 2006 HYSFDFG 2012
> hsa:6790 AURKA, AIK, ARK1, AURA, AURORA2, BTAK, MGC34538, STK15,
STK6, STK7; aurora kinase A (EC:2.7.11.1); K11481 aurora
kinase A [EC:2.7.11.1]
Length=403
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 32/67 (47%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
S+ T + GTL Y PE+++G +D DLWSLGVL +E L G+ PF NT +E KRI
Sbjct 283 SSRRTTLCGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLV-GKPPFEANTYQETYKRI 341
Query 61 IRLEIDF 67
R+E F
Sbjct 342 SRVEFTF 348
> xla:397925 aurka, Eg2, aik, ark1, aura, aurka-a, aurora2, btak,
stk15, stk6, stk7; aurora kinase A (EC:2.7.11.1); K11481
aurora kinase A [EC:2.7.11.1]
Length=407
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 30/72 (41%), Positives = 45/72 (62%), Gaps = 4/72 (5%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
S+ T + GTL Y PE+++G +D + DLWSLGVL +E L G+ PF +T +E +RI
Sbjct 290 SSRRTTLCGTLDYLPPEMIEGRMHDETVDLWSLGVLCYEFLV-GKPPFETDTHQETYRRI 348
Query 61 IRLEIDFEGGPP 72
++E + PP
Sbjct 349 SKVEFQY---PP 357
> xla:398349 aurka-b; p46XlEg22 (EC:2.7.11.1); K11481 aurora kinase
A [EC:2.7.11.1]
Length=408
Score = 60.8 bits (146), Expect = 9e-10, Method: Composition-based stats.
Identities = 30/72 (41%), Positives = 44/72 (61%), Gaps = 4/72 (5%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
S+ T + GTL Y PE+++G +D DLWSLGVL +E L G+ PF +T +E +RI
Sbjct 290 SSRRTTLCGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLV-GKPPFETDTHQETYRRI 348
Query 61 IRLEIDFEGGPP 72
++E + PP
Sbjct 349 SKVEFQY---PP 357
> mmu:20878 Aurka, AIRK1, AU019385, AW539821, Ark1, Aurora-A,
Ayk1, IAK, IAK1, Stk6; aurora kinase A (EC:2.7.11.1); K11481
aurora kinase A [EC:2.7.11.1]
Length=417
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/67 (44%), Positives = 41/67 (61%), Gaps = 1/67 (1%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
S+ T + GTL Y PE+++G +D DLWSLGVL +E L G PF +T +E +RI
Sbjct 296 SSRRTTMCGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLV-GMPPFEAHTYQETYRRI 354
Query 61 IRLEIDF 67
R+E F
Sbjct 355 SRVEFTF 361
> dre:445246 aurka, zgc:100912; aurora kinase A (EC:2.7.11.1);
K11481 aurora kinase A [EC:2.7.11.1]
Length=405
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/67 (41%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
S+ + + GTL Y PE+++G ++D DLWSLGVL +E L GR PF + EE ++I
Sbjct 288 SSRRSTLCGTLDYLPPEMIEGKTHDEKVDLWSLGVLCYEFLV-GRPPFETKSHEETYRKI 346
Query 61 IRLEIDF 67
R+E +
Sbjct 347 SRVEFTY 353
> hsa:6795 AURKC, AIE2, AIK3, ARK3, AurC, STK13, aurora-C; aurora
kinase C (EC:2.7.11.1); K11480 aurora kinase C [EC:2.7.11.1]
Length=309
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE+++G +YD DLW +GVL +ELL G PF + E +RI+++++ F
Sbjct 201 GTLDYLPPEMIEGRTYDEKVDLWCIGVLCYELLV-GYPPFESASHSETYRRILKVDVRFP 259
Query 69 GGPPAG 74
P G
Sbjct 260 LSMPLG 265
> ath:AT3G20830 protein kinase family protein
Length=408
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 43/68 (63%), Gaps = 1/68 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
+ GT Y +PEV++GD +D++ D W+LGVL++E++ G PF G + +E + ++
Sbjct 241 SNSFVGTDEYVSPEVIRGDGHDFAVDWWALGVLTYEMMY-GETPFKGKSKKETFRNVLMK 299
Query 64 EIDFEGGP 71
E +F G P
Sbjct 300 EPEFAGKP 307
> ath:AT2G37840 protein kinase family protein; K08269 unc51-like
kinase [EC:2.7.11.1]
Length=733
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIR- 62
A + G+ Y APE++Q YD ADLWS+G + F+L+T GR PFTGN+ ++ + IIR
Sbjct 168 AETLCGSPLYMAPEIMQLQKYDAKADLWSVGAILFQLVT-GRTPFTGNSQIQLLQNIIRS 226
Query 63 LEIDFEG 69
E+ F G
Sbjct 227 TELHFPG 233
> ath:AT3G53930 protein kinase family protein; K08269 unc51-like
kinase [EC:2.7.11.1]
Length=712
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIR 62
A + G+ Y APE++Q YD ADLWS+G + F+L+T GR PFTGN+ ++ + IIR
Sbjct 176 AETLCGSPLYMAPEIMQLQKYDAKADLWSVGAILFQLVT-GRTPFTGNSQIQLLQNIIR 233
> ath:AT3G04530 PPCK2; PPCK2; kinase/ protein serine/threonine
kinase
Length=278
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 41/69 (59%), Gaps = 4/69 (5%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A V GT Y APEVV G YD D+WS GV+ + +L +G PF G TAE++ + I+R
Sbjct 168 AEGVVGTPYYVAPEVVMGRKYDEKVDIWSAGVVIYTML-AGEPPFNGETAEDIFESILRG 226
Query 64 EIDFEGGPP 72
+ F PP
Sbjct 227 NLRF---PP 232
> mmu:20871 Aurkc, AIE1, AIK3, IAK3, Stk13; aurora kinase C (EC:2.7.11.1);
K11480 aurora kinase C [EC:2.7.11.1]
Length=315
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE++ Y+ DLW +GVL +ELL G+ PF +T+ E +RI +++ F
Sbjct 213 GTLDYLPPEMIAQKPYNEMVDLWCIGVLCYELLV-GKPPFESSTSSETYRRIRQVDFKFP 271
Query 69 GGPPAG 74
PAG
Sbjct 272 SSVPAG 277
> ath:AT1G51170 protein kinase family protein
Length=404
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 42/68 (61%), Gaps = 1/68 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
+ GT Y +PEV++GD +D++ D W+LGVL++E++ G PF G +E + ++
Sbjct 240 SNSFVGTDEYISPEVIRGDGHDFAVDWWALGVLTYEMMY-GETPFKGRNKKETFRNVLVK 298
Query 64 EIDFEGGP 71
E +F G P
Sbjct 299 EPEFAGKP 306
> sce:YPL209C IPL1, PAC15; Aurora kinase subunit of the conserved
chromosomal passenger complex (CPC; Ipl1p-Sli15p-Bir1p-Nbl1p),
involved in regulating kinetochore-microtubule attachments;
helps maintain condensed chromosomes during anaphase
and early telophase (EC:2.7.11.1); K08850 aurora kinase, other
[EC:2.7.11.1]
Length=367
Score = 57.8 bits (138), Expect = 7e-09, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 1/59 (1%)
Query 7 VTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEI 65
V GT+ Y +PE+V+ YD++ D W+LGVL+FELLT G PF + KRI L+I
Sbjct 261 VCGTIDYLSPEMVESREYDHTIDAWALGVLAFELLT-GAPPFEEEMKDTTYKRIAALDI 318
> ath:AT2G45490 AtAUR3; AtAUR3 (ATAURORA3); ATP binding / histone
kinase(H3-S10 specific) / protein kinase; K08850 aurora
kinase, other [EC:2.7.11.1]
Length=288
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y APE+V+ +DY+ D W+LG+L +E L G PF + ++ KRI+++++ F
Sbjct 179 GTLDYLAPEMVENRDHDYAVDNWTLGILCYEFLY-GNPPFEAESQKDTFKRILKIDLSFP 237
Query 69 GGP 71
P
Sbjct 238 LTP 240
> hsa:9212 AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, STK12,
STK5, aurkb-sv1, aurkb-sv2; aurora kinase B (EC:2.7.11.1); K11479
aurora kinase B [EC:2.7.11.1]
Length=344
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE+++G ++ DLW +GVL +ELL G PF + E +RI+++++ F
Sbjct 235 GTLDYLPPEMIEGRMHNEKVDLWCIGVLCYELLV-GNPPFESASHNETYRRIVKVDLKFP 293
Query 69 GGPPAG 74
P G
Sbjct 294 ASVPMG 299
> dre:791894 aurkb, CHUNP6896, MGC85918, fa09g06, stka, wu:fa09g06;
aurora kinase B (EC:2.7.11.1); K11479 aurora kinase B
[EC:2.7.11.1]
Length=320
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDF 67
GTL Y PE+++G S+D DLWS+GVL +E L G PF + E KRI ++++ F
Sbjct 211 GTLDYLPPEMIEGHSHDEKVDLWSIGVLCYECLV-GNPPFETASHAETYKRITKVDLQF 268
> mmu:209012 Ulk4, 4932415A06Rik, A730098P15; unc-51-like kinase
4 (C. elegans) (EC:2.7.11.1)
Length=1275
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 5 TEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
T V G+L YAAPE+V+G + ++DLWSLG L +E+ SG+ PF T E+ ++I+
Sbjct 177 TRVRGSLIYAAPEIVKGTEFSVTSDLWSLGCLLYEMF-SGKPPFFSETVSELVEKIL 232
> cel:F17E5.1 lin-2; abnormal cell LINeage family member (lin-2);
K06103 calcium/calmodulin-dependent serine protein kinase
[EC:2.7.11.1]
Length=961
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 45/64 (70%), Gaps = 2/64 (3%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
G + APE+V+ D S+D+WS GV+ F LL +GRLPF+G+T+ ++ +RI++ ++D +
Sbjct 187 GVPQFMAPEIVRKDRVSCSSDIWSSGVVLF-LLLAGRLPFSGSTS-DIYERIMQTDVDVD 244
Query 69 GGPP 72
G P
Sbjct 245 GYMP 248
> hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protein
S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1); K04445 mitogen-,stress
activated protein kinases [EC:2.7.11.1]
Length=802
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/67 (44%), Positives = 43/67 (64%), Gaps = 8/67 (11%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPF-------TGNTAEEVKKRIIR 62
TL YAAPE++ + YD S DLWSLGV+ + +L SG++PF T +A E+ K+I +
Sbjct 585 TLHYAAPELLNQNGYDESCDLWSLGVILYTML-SGQVPFQSHDRSLTCTSAVEIMKKIKK 643
Query 63 LEIDFEG 69
+ FEG
Sbjct 644 GDFSFEG 650
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 43/67 (64%), Gaps = 7/67 (10%)
Query 4 ATEVTGTLSYAAPEVVQG-DS-YDYSADLWSLGVLSFELLTSGRLPFT----GNTAEEVK 57
A GT+ Y AP++V+G DS +D + D WSLGVL +ELLT G PFT N+ E+
Sbjct 210 AYSFCGTIEYMAPDIVRGGDSGHDKAVDWWSLGVLMYELLT-GASPFTVDGEKNSQAEIS 268
Query 58 KRIIRLE 64
+RI++ E
Sbjct 269 RRILKSE 275
> dre:565173 ribosomal protein S6 kinase, polypeptide 4-like;
K04445 mitogen-,stress activated protein kinases [EC:2.7.11.1]
Length=811
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/67 (43%), Positives = 43/67 (64%), Gaps = 8/67 (11%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPF-------TGNTAEEVKKRIIR 62
TL YAAPE+++ YD S DLWSLGV+ + +L SG++PF +AEE+ ++I +
Sbjct 581 TLQYAAPEILKYSGYDESCDLWSLGVILYTML-SGQVPFQCQGKSLMHTSAEEIMRKIKQ 639
Query 63 LEIDFEG 69
+ FEG
Sbjct 640 GDFSFEG 646
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 46/78 (58%), Gaps = 9/78 (11%)
Query 4 ATEVTGTLSYAAPEVVQGDS--YDYSADLWSLGVLSFELLTSGRLPFT----GNTAEEVK 57
A + GT+ Y APE+V G +D + D WS+GVL +ELLT G PFT N+ ++
Sbjct 206 AYSICGTIEYMAPEIVAGGEGGHDKAVDWWSMGVLMYELLTGGS-PFTVDGDENSHSDIA 264
Query 58 KRIIRLEIDF--EGGPPA 73
+RI++ + F + GP A
Sbjct 265 ERIMKKDPPFPKDMGPLA 282
> dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=3712
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 39/59 (66%), Gaps = 1/59 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDF 67
GTL Y +PE+++GD AD+WS+G+L++ +L SGRLPFT N E + RI + D
Sbjct 3561 GTLDYMSPEMLKGDVVGPPADIWSIGILTYIML-SGRLPFTENDPAETEARIQAAKFDL 3618
> mmu:628475 Gm6882, EG628475; predicted gene 6882
Length=313
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 38/65 (58%), Gaps = 2/65 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDF 67
GTL Y APE++Q + Y+ D+WSLGVL F L+ SG LPF G + E+K+ II
Sbjct 189 GTLPYCAPELLQAEKYEGLPVDIWSLGVLLF-LMVSGNLPFQGRSFVELKQEIISANFSI 247
Query 68 EGGPP 72
P
Sbjct 248 PSNVP 252
> mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC28385,
MSK1, MSPK1, RLPK; ribosomal protein S6 kinase, polypeptide
5 (EC:2.7.11.1); K04445 mitogen-,stress activated protein
kinases [EC:2.7.11.1]
Length=863
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/67 (44%), Positives = 43/67 (64%), Gaps = 8/67 (11%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPF-------TGNTAEEVKKRIIR 62
TL YAAPE++ + YD S DLWSLGV+ + +L SG++PF T +A E+ K+I +
Sbjct 649 TLHYAAPELLTHNGYDESCDLWSLGVILYTML-SGQVPFQSHDRSLTCTSAVEIMKKIKK 707
Query 63 LEIDFEG 69
+ FEG
Sbjct 708 GDFSFEG 714
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 43/67 (64%), Gaps = 7/67 (10%)
Query 4 ATEVTGTLSYAAPEVVQG-DS-YDYSADLWSLGVLSFELLTSGRLPFT----GNTAEEVK 57
A GT+ Y AP++V+G DS +D + D WSLGVL +ELLT G PFT N+ E+
Sbjct 209 AYSFCGTIEYMAPDIVRGGDSGHDKAVDWWSLGVLMYELLT-GASPFTVDGEKNSQAEIS 267
Query 58 KRIIRLE 64
+RI++ E
Sbjct 268 RRILKSE 274
> dre:561959 si:dkey-273o13.5 (EC:2.7.11.1); K04445 mitogen-,stress
activated protein kinases [EC:2.7.11.1]
Length=745
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 35/72 (48%), Positives = 44/72 (61%), Gaps = 8/72 (11%)
Query 9 GTLSYAAPEVVQGDS-YDYSADLWSLGVLSFELLTSGRLPFT----GNTAEEVKKRIIRL 63
GT+ Y APE+++G S + + D WSLG+L FELLT G PFT N+ EV KRI+R
Sbjct 170 GTIEYMAPEIIRGKSGHGKAVDWWSLGILMFELLT-GASPFTLEGERNSQSEVSKRILRC 228
Query 64 EIDFEG--GPPA 73
E F GP A
Sbjct 229 EPPFPSIIGPLA 240
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/39 (53%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPF 48
TL YAAPE+ YD + DLWSLGV+ + +L SG++PF
Sbjct 541 TLQYAAPELFHSSGYDQACDLWSLGVILYTML-SGQVPF 578
> mmu:20877 Aurkb, AIM-1, AIRK2, AL022959, Aik2, Aim1, Ark2, AurB,
IPL1, STK-1, Stk12, Stk5; aurora kinase B (EC:2.7.11.1);
K11479 aurora kinase B [EC:2.7.11.1]
Length=345
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE+++G ++ DLW +GVL +EL+ G PF + E +RI+++++ F
Sbjct 240 GTLDYLPPEMIEGRMHNEMVDLWCIGVLCYELMV-GNPPFESPSHSETYRRIVKVDLKFP 298
Query 69 GGPPAG 74
P+G
Sbjct 299 SSVPSG 304
> ath:AT3G08730 PK1; PK1 (PROTEIN-SERINE KINASE 1); kinase/ protein
binding / protein kinase/ protein serine/threonine kinase;
K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=465
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 46/65 (70%), Gaps = 2/65 (3%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
+T + + GT Y APE+V+G +D +AD WS+G+L +E+LT G+ PF G+ +++++I
Sbjct 285 NTRSNSMCGTTEYMAPEIVRGKGHDKAADWWSVGILLYEMLT-GKPPFLGSKG-KIQQKI 342
Query 61 IRLEI 65
++ +I
Sbjct 343 VKDKI 347
> dre:557667 trio; si:dkey-158b13.2 (EC:2.7.11.1); K08810 triple
functional domain protein [EC:2.7.11.1]
Length=3087
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDF 67
G+ +AAPE+V GD S+DLWSLGVL++ +L SG PF + EE I RL+ F
Sbjct 2943 GSPEFAAPELVLGDPVSLSSDLWSLGVLTYVML-SGASPFLDESVEETCLNICRLDFSF 3000
> dre:337670 rps6kal, fj94b02, rps6ka6, wu:fj94b02, zgc:66139;
ribosomal protein S6 kinase, like (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=740
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 29/62 (46%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + SAD WSLGVL FE+LT G LPF G E I++
Sbjct 224 AYSFCGTVEYMAPEVVNRRGHTQSADWWSLGVLMFEMLT-GTLPFQGKDRNETMNMILKA 282
Query 64 EI 65
++
Sbjct 283 KL 284
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 36/64 (56%), Gaps = 4/64 (6%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFT---GNTAEEVKKRIIRLEID 66
T ++ APEV+ YD + D+WSLGVL + +L +G PF +T EE+ RI +
Sbjct 579 TANFVAPEVLMRQGYDAACDIWSLGVLLYTML-AGYTPFANGPNDTPEEILLRIGSGKFS 637
Query 67 FEGG 70
GG
Sbjct 638 LSGG 641
> ath:AT2G25880 AtAUR2; AtAUR2 (AtAurora2); histone kinase(H3-S10
specific) / kinase; K08850 aurora kinase, other [EC:2.7.11.1]
Length=256
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE+V+ +D S D+WSLG+L +E L G PF E KRI+++++ F
Sbjct 150 GTLDYLPPEMVESVEHDASVDIWSLGILCYEFLY-GVPPFEAREHSETYKRIVQVDLKFP 208
Query 69 GGP 71
P
Sbjct 209 PKP 211
> hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protein
S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=744
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + +SAD WS GVL FE+LT G LPF G +E I++
Sbjct 228 AYSFCGTVEYMAPEVVNRQGHSHSADWWSYGVLMFEMLT-GSLPFQGKDRKETMTLILKA 286
Query 64 EI 65
++
Sbjct 287 KL 288
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 4/54 (7%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTG---NTAEEVKKRI 60
T ++ APEV++ YD D+WSLG+L + +L +G PF +T EE+ RI
Sbjct 586 TANFVAPEVLKRQGYDEGCDIWSLGILLYTML-AGYTPFANGPSDTPEEILTRI 638
> cpv:cgd3_260 hypothetical protein
Length=1180
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/59 (42%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDF 67
GTL Y APE++ G YD D+WS+GV++ +L G PF GNT E+ +++ R + F
Sbjct 388 GTLYYIAPEILLGKGYDDKCDIWSIGVMA-HILLCGVPPFAGNTDTEIIEKVRRGSVSF 445
> ath:AT3G08720 S6K2; S6K2 (ARABIDOPSIS THALIANA SERINE/THREONINE
PROTEIN KINASE 2); kinase/ protein kinase
Length=471
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 46/65 (70%), Gaps = 2/65 (3%)
Query 1 STGATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRI 60
+T + + GT Y APE+V+G +D +AD WS+G+L +E+LT G+ PF G+ +++++I
Sbjct 291 NTRSNSMCGTTEYMAPEIVRGKGHDKAADWWSVGILLYEMLT-GKPPFLGSKG-KIQQKI 348
Query 61 IRLEI 65
++ +I
Sbjct 349 VKDKI 353
> xla:443729 rps6ka6, MGC79981, rps6ka, rps6ka1; ribosomal protein
S6 kinase, 90kDa, polypeptide 6 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + +SAD WS GVL FE+LT G LPF G +E I++
Sbjct 219 AYSFCGTVEYMAPEVVNRQGHSHSADWWSYGVLMFEMLT-GSLPFQGKDRKETMTLILKA 277
Query 64 EI 65
++
Sbjct 278 KL 279
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 4/54 (7%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFT---GNTAEEVKKRI 60
T ++ APEV++ YD D+WSLG+L + +L +G PF G+T EE+ RI
Sbjct 575 TANFVAPEVLKRQGYDEGCDIWSLGILLYTML-AGYTPFANGLGDTPEEILARI 627
> dre:100009657 rps6ka3b, zgc:158386; ribosomal protein S6 kinase,
polypeptide 3b (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=720
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + YSAD WS GVL +E+LT G LPF G +E I++
Sbjct 224 AYSFCGTVEYMAPEVVNRRGHTYSADWWSYGVLMYEMLT-GTLPFQGKDRKETMTMILKA 282
Query 64 EI 65
++
Sbjct 283 KL 284
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/64 (40%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTG---NTAEEVKKRIIRLEID 66
T ++ APEV++ YD + D+WSLGVL + +LT G PF +T EE+ RI +
Sbjct 550 TANFVAPEVLKKQGYDAACDIWSLGVLLYTMLT-GFTPFANGPEDTPEEILARIGSGQFS 608
Query 67 FEGG 70
GG
Sbjct 609 LTGG 612
> cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog family
member (rskn-1); K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=804
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 1/57 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEI 65
GT+ Y APEV+ + +AD WSLGVL FE+LT G LPF G + +I++ ++
Sbjct 260 GTVEYMAPEVINRRGHSMAADFWSLGVLMFEMLT-GHLPFQGRDRNDTMTQILKAKL 315
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 4/54 (7%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFT---GNTAEEVKKRI 60
T + APEV++ YD S D+WSLGVL +LT G PF +T +++ +R+
Sbjct 608 TAQFVAPEVLRKQGYDRSCDVWSLGVLLHTMLT-GCTPFAMGPNDTPDQILQRV 660
> mmu:100043665 Gm10662; predicted gene 10662
Length=313
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 27/54 (50%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
GTL Y APE++Q + Y+ D+WSLGVL F L+ SG LPF G + ++K+ II
Sbjct 189 GTLPYCAPELLQAEKYEGLPVDIWSLGVLLF-LMVSGNLPFQGRSFVDLKQEII 241
> mmu:545929 Gm5891, EG545929; predicted gene 5891
Length=313
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 27/54 (50%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
GTL Y APE++Q + Y+ D+WSLGVL F L+ SG LPF G + ++K+ II
Sbjct 189 GTLPYCAPELLQAEKYEGLPVDIWSLGVLLF-LMVSGNLPFQGRSFVDLKQEII 241
> mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kinase
polypeptide 1 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=735
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + +SAD WS GVL FE+LT G LPF G +E I++
Sbjct 219 AYSFCGTVEYMAPEVVNRQGHTHSADWWSYGVLMFEMLT-GSLPFQGKDRKETMTLILKA 277
Query 64 EI 65
++
Sbjct 278 KL 279
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 4/54 (7%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTG---NTAEEVKKRI 60
T ++ APEV++ YD D+WSLG+L + +L +G PF +T EE+ RI
Sbjct 577 TANFVAPEVLKRQGYDEGCDIWSLGILLYTML-AGYTPFANGPSDTPEEILTRI 629
> hsa:57787 MARK4, FLJ12177, FLJ90097, KIAA1860, MARK4L, MARK4S,
MARKL1, MARKL1L; MAP/microtubule affinity-regulating kinase
4 (EC:2.7.11.1); K08798 MAP/microtubule affinity-regulating
kinase [EC:2.7.11.1]
Length=752
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 39/56 (69%), Gaps = 2/56 (3%)
Query 8 TGTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIR 62
G+ YAAPE+ QG YD D+WSLGV+ + L+ SG LPF G+ +E+++R++R
Sbjct 216 CGSPPYAAPELFQGKKYDGPEVDIWSLGVILYTLV-SGSLPFDGHNLKELRERVLR 270
> dre:100330808 MAP/microtubule affinity-regulating kinase 3-like
Length=527
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 26/55 (47%), Positives = 38/55 (69%), Gaps = 2/55 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIR 62
G+ YAAPE+ QG YD D+WSLGV+ + L+ SG LPF G +E+++R++R
Sbjct 216 GSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLV-SGSLPFDGQNLKELRERVLR 269
> mmu:232944 Mark4, 2410090P21Rik, C79806, Markl1; MAP/microtubule
affinity-regulating kinase 4 (EC:2.7.11.1); K08798 MAP/microtubule
affinity-regulating kinase [EC:2.7.11.1]
Length=752
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 39/56 (69%), Gaps = 2/56 (3%)
Query 8 TGTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIR 62
G+ YAAPE+ QG YD D+WSLGV+ + L+ SG LPF G+ +E+++R++R
Sbjct 216 CGSPPYAAPELFQGKKYDGPEVDIWSLGVILYTLV-SGSLPFDGHNLKELRERVLR 270
> dre:555536 si:dkey-240h12.4; K08803 death-associated protein
kinase [EC:2.7.11.1]
Length=358
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 38/60 (63%), Gaps = 1/60 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GT Y APE++ + +AD+WS+GV+++ LL SG PF G T EE + I+ + +FE
Sbjct 179 GTPQYIAPEIINYEPLGTAADMWSIGVITYILL-SGLSPFQGETDEETLRNIVSMNYEFE 237
> ath:AT3G25250 AGC2-1; AGC2-1 (OXIDATIVE SIGNAL-INDUCIBLE1);
kinase
Length=421
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 25/53 (47%), Positives = 35/53 (66%), Gaps = 1/53 (1%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
GT Y APEV+ GD +D++ D WSLGV+ +E+L G PF G+ +E RI+
Sbjct 238 GTEEYVAPEVISGDGHDFAVDWWSLGVVLYEMLY-GATPFRGSNRKETFYRIL 289
> dre:571285 ribosomal protein S6 kinase, polypeptide 2-like;
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + SAD WS GVL FE+LT G LPF G +E I++
Sbjct 216 AYSFCGTIEYMAPEVVNRRGHTQSADWWSFGVLMFEMLT-GSLPFQGKDRKETMALILKA 274
Query 64 EI 65
++
Sbjct 275 KL 276
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTG---NTAEEVKKRIIRLEID 66
T ++ APEV++ YD + D+WSLG+L + +L +G PF +T EE+ RI +
Sbjct 574 TANFVAPEVLKKQGYDAACDIWSLGILLYTML-AGFTPFANGPDDTPEEILARIGSGKFA 632
Query 67 FEGG 70
GG
Sbjct 633 LSGG 636
> mmu:100043625 Gm4557; predicted gene 4557
Length=306
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/54 (50%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
GTL Y APE++Q + Y+ D+WSLGVL F L+ SG LPF G + ++K+ II
Sbjct 182 GTLPYCAPELLQAEKYEGPPVDIWSLGVLLF-LMVSGNLPFQGKSFVDLKQEII 234
> mmu:545926 Gm5890, EG545926; predicted gene 5890
Length=306
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/54 (50%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 9 GTLSYAAPEVVQGDSYD-YSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRII 61
GTL Y APE++Q + Y+ D+WSLGVL F L+ SG LPF G + ++K+ II
Sbjct 182 GTLPYCAPELLQAEKYEGPPVDIWSLGVLLF-LMVSGNLPFQGKSFVDLKQEII 234
> hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-alpha2,
p90-RSK3, pp90RSK3; ribosomal protein S6 kinase, 90kDa,
polypeptide 2 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=741
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + SAD WS GVL FE+LT G LPF G +E I++
Sbjct 224 AYSFCGTIEYMAPEVVNRRGHTQSADWWSFGVLMFEMLT-GSLPFQGKDRKETMALILKA 282
Query 64 EI 65
++
Sbjct 283 KL 284
Score = 44.3 bits (103), Expect = 8e-05, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query 10 TLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTG---NTAEEVKKRIIRLEID 66
T ++ APEV++ YD + D+WSLG+L + +L +G PF +T EE+ RI +
Sbjct 582 TANFVAPEVLKRQGYDAACDIWSLGILLYTML-AGFTPFANGPDDTPEEILARIGSGKYA 640
Query 67 FEGG 70
GG
Sbjct 641 LSGG 644
> xla:444083 aurkb-b, AurB, MGC83575, aik2, aim1, airk2, ark2,
ipl1, stk12, stk5; aurora kinase B (EC:2.7.11.1); K11479 aurora
kinase B [EC:2.7.11.1]
Length=368
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query 9 GTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRLEIDFE 68
GTL Y PE+++G ++D DLW GVL +E L G PF + E +RI+ +++ F
Sbjct 258 GTLDYLPPEMIEGKTHDEKVDLWCAGVLCYEFLV-GMPPFDSPSHSETHRRIVNVDLKF- 315
Query 69 GGPP 72
PP
Sbjct 316 --PP 317
> mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk,
pp90rsk; ribosomal protein S6 kinase, polypeptide 2 (EC:2.7.11.1);
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 4 ATEVTGTLSYAAPEVVQGDSYDYSADLWSLGVLSFELLTSGRLPFTGNTAEEVKKRIIRL 63
A GT+ Y APEVV + SAD WS GVL FE+LT G LPF G +E I++
Sbjct 216 AYSFCGTIEYMAPEVVNRRGHTQSADWWSFGVLMFEMLT-GSLPFQGKDRKETMALILKA 274
Query 64 EI 65
++
Sbjct 275 KL 276
Lambda K H
0.312 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007182052
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40