bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8167_orf1
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
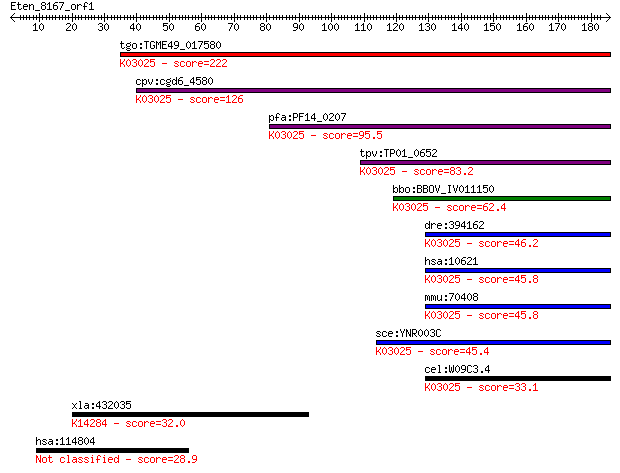
tgo:TGME49_017580 DNA-directed RNA polymerases III, 39 kDa pol... 222 6e-58
cpv:cgd6_4580 RNA polymerase III C34 subunit; rpc34p ortholog ... 126 5e-29
pfa:PF14_0207 RNA polymerase subunit, putative; K03025 DNA-dir... 95.5 1e-19
tpv:TP01_0652 RNA polymerase subunit; K03025 DNA-directed RNA ... 83.2 4e-16
bbo:BBOV_IV011150 23.m06053; RNA polymerase rpc34 subunit cont... 62.4 8e-10
dre:394162 polr3f, MGC56299, zgc:56299, zgc:76983; polymerase ... 46.2 7e-05
hsa:10621 POLR3F, MGC13517, RPC39, RPC6; polymerase (RNA) III ... 45.8 9e-05
mmu:70408 Polr3f, 2810411G20Rik, 3010019O03Rik, 3110032A07Rik,... 45.8 9e-05
sce:YNR003C RPC34; C34; K03025 DNA-directed RNA polymerase III... 45.4 1e-04
cel:W09C3.4 hypothetical protein; K03025 DNA-directed RNA poly... 33.1 0.51
xla:432035 nxf1, MGC82224; nuclear RNA export factor 1; K14284... 32.0 1.4
hsa:114804 RNF157; ring finger protein 157 28.9
> tgo:TGME49_017580 DNA-directed RNA polymerases III, 39 kDa polypeptide,
putative ; K03025 DNA-directed RNA polymerase III
subunit RPC6
Length=325
Score = 222 bits (565), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 102/151 (67%), Positives = 121/151 (80%), Gaps = 0/151 (0%)
Query 35 TPRGAPAGAPRGSSALSGEDLSTAYNLGLRNNNELTQQLLLDQGWPREKIVAVFNRLNEA 94
+P+ P+ S+ L+ DL AY LG +++NELTQ+LLL QGW +EKIVA FNRL EA
Sbjct 11 SPQALPSPQASTSATLTATDLQAAYTLGQQHDNELTQELLLQQGWGKEKIVAAFNRLTEA 70
Query 95 RAGVIRKKGDNICCVVRPANVIPALKQLDALDYKVYCAVELAGNTGVWTADIRKSTGLQT 154
RA VIR+KG +CC++RP+NVI LK LDA DYKVYCA+E AG TGVWTAD+RKSTGLQT
Sbjct 71 RAAVIRRKGTGLCCLLRPSNVIGKLKTLDAFDYKVYCAIEQAGTTGVWTADLRKSTGLQT 130
Query 155 HIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
HIIQR VK LC+ L+LIKPVKSIHVKNRKMY
Sbjct 131 HIIQRSVKQLCDFLKLIKPVKSIHVKNRKMY 161
> cpv:cgd6_4580 RNA polymerase III C34 subunit; rpc34p ortholog
; K03025 DNA-directed RNA polymerase III subunit RPC6
Length=319
Score = 126 bits (316), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 67/155 (43%), Positives = 90/155 (58%), Gaps = 9/155 (5%)
Query 40 PAGAPRGSSALSGEDLSTAYNLGLRNNNELTQQLLLDQGWPREKIVAVFNRLNEAR---- 95
P GA + ++ L AY G +NNNEL+ L GW IV V N E R
Sbjct 4 PGGAGGSKTQITAAILQEAYIHGCQNNNELSPDSLKSLGWDNSVIVRVLNIFTEKRLCSV 63
Query 96 -----AGVIRKKGDNICCVVRPANVIPALKQLDALDYKVYCAVELAGNTGVWTADIRKST 150
+G++ + + +R V L +L +Y V+C++E AGN G+WTADIRK+T
Sbjct 64 KKNRPSGLLSSVNNKVVFQLRTEEVACKLNKLSNEEYLVFCSIEDAGNQGIWTADIRKNT 123
Query 151 GLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
GLQTH++QR VK LC + LI+PVKSIHVKNRK+Y
Sbjct 124 GLQTHVVQRAVKVLCNDWNLIRPVKSIHVKNRKVY 158
> pfa:PF14_0207 RNA polymerase subunit, putative; K03025 DNA-directed
RNA polymerase III subunit RPC6
Length=311
Score = 95.5 bits (236), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 50/105 (47%), Positives = 67/105 (63%), Gaps = 1/105 (0%)
Query 81 REKIVAVFNRLNEARAGVIRKKGDNICCVVRPANVIPALKQLDALDYKVYCAVELAGNTG 140
R +IV N L ARA I+ + + I +R V LK+L +D+ ++ VE + N G
Sbjct 42 RNEIVYALNILENARACSIKNENNTIITRMRNEQVTKKLKELSDIDFLIFTKVENSQNNG 101
Query 141 VWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
+WTAD+RK T L H +Q+GVK LCEN +LIK V +IHVKNRKMY
Sbjct 102 IWTADLRKQTKLLIHQVQKGVKLLCEN-KLIKQVNNIHVKNRKMY 145
> tpv:TP01_0652 RNA polymerase subunit; K03025 DNA-directed RNA
polymerase III subunit RPC6
Length=295
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 0/77 (0%)
Query 109 VVRPANVIPALKQLDALDYKVYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENL 168
V+R +++ L +L +DYK++C +E AGN G+WTADIRK GL + +QRGVK L E+
Sbjct 74 VLRDDDLVGKLMKLKDIDYKIFCTIETAGNKGIWTADIRKILGLHINQVQRGVKYLYESN 133
Query 169 QLIKPVKSIHVKNRKMY 185
+LI+P+ ++ K+RK++
Sbjct 134 RLIRPITDVNYKSRKLF 150
> bbo:BBOV_IV011150 23.m06053; RNA polymerase rpc34 subunit containing
protein; K03025 DNA-directed RNA polymerase III subunit
RPC6
Length=293
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 119 LKQLDALDYKVYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIH 178
+ +L L+Y V+CA+E AG+ G+WTADI+K + + + R +K L E L+K V ++H
Sbjct 85 VSRLHDLEYAVFCAIETAGDRGIWTADIKKICEITGNQLTRSLKVLLEQHGLVKQVTNVH 144
Query 179 VKNRKMY 185
K+RK+Y
Sbjct 145 QKSRKLY 151
> dre:394162 polr3f, MGC56299, zgc:56299, zgc:76983; polymerase
(RNA) III (DNA directed) polypeptide F; K03025 DNA-directed
RNA polymerase III subunit RPC6
Length=314
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 1/57 (1%)
Query 129 VYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
VY +E AGN G+W+ DIR + L I + +KNL E+ +LIK VKS+ +K+Y
Sbjct 93 VYQIIEDAGNKGIWSRDIRYKSNLPLTEINKILKNL-ESKKLIKAVKSVAASKKKVY 148
> hsa:10621 POLR3F, MGC13517, RPC39, RPC6; polymerase (RNA) III
(DNA directed) polypeptide F, 39 kDa; K03025 DNA-directed
RNA polymerase III subunit RPC6
Length=316
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 1/57 (1%)
Query 129 VYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
VY +E AGN G+W+ DIR + L I + +KNL E+ +LIK VKS+ +K+Y
Sbjct 95 VYQIIEDAGNKGIWSRDIRYKSNLPLTEINKILKNL-ESKKLIKAVKSVAASKKKVY 150
> mmu:70408 Polr3f, 2810411G20Rik, 3010019O03Rik, 3110032A07Rik,
MGC11656, RPC39, RPC6; polymerase (RNA) III (DNA directed)
polypeptide F; K03025 DNA-directed RNA polymerase III subunit
RPC6
Length=316
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 1/57 (1%)
Query 129 VYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
VY +E AGN G+W+ DIR + L I + +KNL E+ +LIK VKS+ +K+Y
Sbjct 95 VYQIIEDAGNKGIWSRDIRYKSNLPLTEINKILKNL-ESKKLIKAVKSVAASKKKVY 150
> sce:YNR003C RPC34; C34; K03025 DNA-directed RNA polymerase III
subunit RPC6
Length=317
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 45/91 (49%), Gaps = 20/91 (21%)
Query 114 NVIPALKQLDALDYK-------------------VYCAVELAGNTGVWTADIRKSTGLQT 154
N+I +KQ D L ++ VY +E +G G+W+ I+ T L
Sbjct 59 NLIKLVKQNDELKFQGVLESEAQKKATMSAEEALVYSYIEASGREGIWSKTIKARTNLHQ 118
Query 155 HIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
H++ + +K+L E+ + +K VKS+ RK+Y
Sbjct 119 HVVLKCLKSL-ESQRYVKSVKSVKFPTRKIY 148
> cel:W09C3.4 hypothetical protein; K03025 DNA-directed RNA polymerase
III subunit RPC6
Length=296
Score = 33.1 bits (74), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Query 129 VYCAVELAGNTGVWTADIRKSTGLQTHIIQRGVKNLCENLQLIKPVKSIHVKNRKMY 185
+Y +E + G+W ++R+ +GL +++ +K+L E +LIK +K++ RK Y
Sbjct 88 IYSLIEESKTRGIWIKELREGSGLNQLQLRKTLKSL-ETKKLIKTIKAVGT-TRKCY 142
> xla:432035 nxf1, MGC82224; nuclear RNA export factor 1; K14284
nuclear RNA export factor 1/2
Length=614
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 32/73 (43%), Gaps = 6/73 (8%)
Query 20 ATGEPKDEGARATAETPRGAPAGAPRGSSALSGEDLSTAYNLGLRNNNELTQQLLLDQGW 79
A E + TA TP +P L+ +D+ A+ N+E + + L D GW
Sbjct 536 ANTEEIHKAFATTAPTPSSSPV------HVLASQDMVLAFIQLSGMNSEWSHKCLEDNGW 589
Query 80 PREKIVAVFNRLN 92
++ VF +LN
Sbjct 590 DYDQATQVFMKLN 602
> hsa:114804 RNF157; ring finger protein 157
Length=679
Score = 28.9 bits (63), Expect = 9.2, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 22/47 (46%), Gaps = 2/47 (4%)
Query 9 GSRMSAPKEGSATGEPKDEGARATAETPRGAPAGAPRGSSALSGEDL 55
G +S+P+ S P +EG AE+P AG P G G D+
Sbjct 548 GEALSSPQPASRA--PSEEGEGLPAESPDSNFAGLPAGEQDAEGNDV 592
Lambda K H
0.315 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5106150148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40