bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8140_orf1
Length=126
Score E
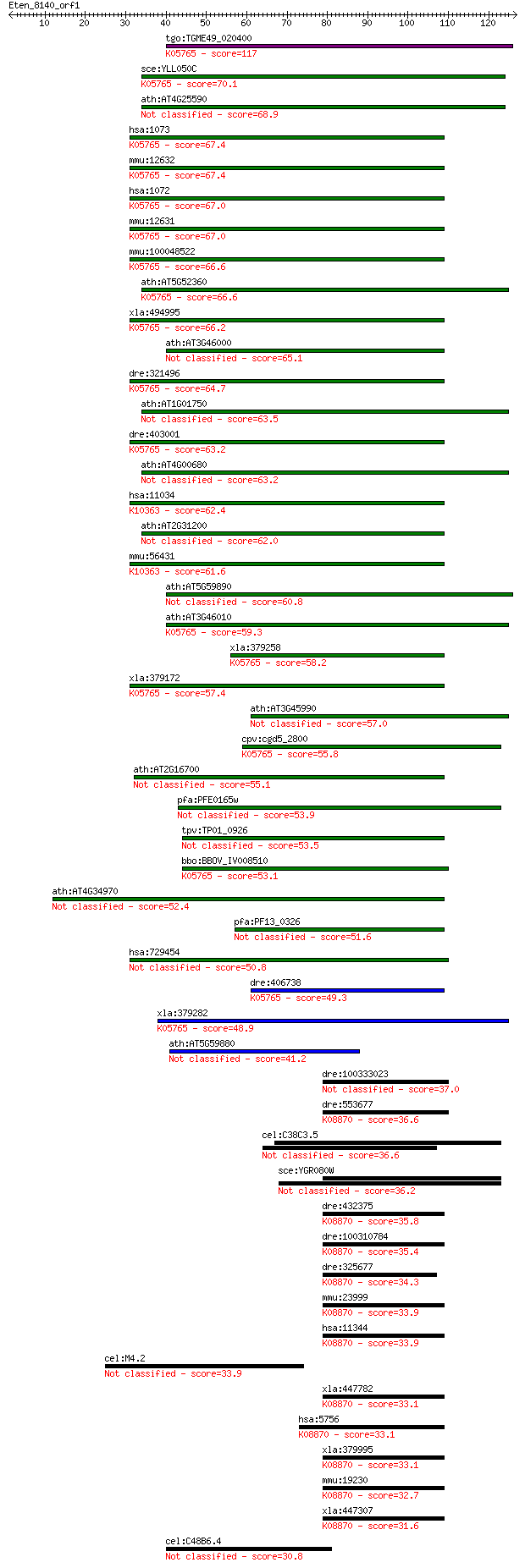
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_020400 actin depolymerizing factor ; K05765 cofilin 117 1e-26
sce:YLL050C COF1; Cof1p; K05765 cofilin 70.1 2e-12
ath:AT4G25590 ADF7; ADF7 (actin depolymerizing factor 7); acti... 68.9 4e-12
hsa:1073 CFL2, NEM7; cofilin 2 (muscle); K05765 cofilin 67.4 1e-11
mmu:12632 Cfl2; cofilin 2, muscle; K05765 cofilin 67.4 1e-11
hsa:1072 CFL1, CFL; cofilin 1 (non-muscle); K05765 cofilin 67.0 1e-11
mmu:12631 Cfl1, AA959946, Cof; cofilin 1, non-muscle; K05765 c... 67.0 2e-11
mmu:100048522 cofilin-1-like; K05765 cofilin 66.6 2e-11
ath:AT5G52360 ADF10; ADF10 (ACTIN DEPOLYMERIZING FACTOR 10); a... 66.6 2e-11
xla:494995 cfl2; cofilin 2 (non-muscle); K05765 cofilin 66.2 3e-11
ath:AT3G46000 ADF2; ADF2 (ACTIN DEPOLYMERIZING FACTOR 2); acti... 65.1 5e-11
dre:321496 cfl2l, CFL2, wu:fb17d06, wu:fb18d11, wu:fd59f08, wu... 64.7 6e-11
ath:AT1G01750 ADF11; ADF11 (ACTIN DEPOLYMERIZING FACTOR 11); a... 63.5 2e-10
dre:403001 cfl2, MGC77288, zgc:77288; cofilin 2 (muscle); K057... 63.2 2e-10
ath:AT4G00680 ADF8; ADF8 (ACTIN DEPOLYMERIZING FACTOR 8); acti... 63.2 2e-10
hsa:11034 DSTN, ACTDP, ADF, bA462D18.2; destrin (actin depolym... 62.4 4e-10
ath:AT2G31200 ADF6; ADF6 (ACTIN DEPOLYMERIZING FACTOR 6); acti... 62.0 4e-10
mmu:56431 Dstn, 2610043P17Rik, ADF, AU042046, Dsn, corn1, sid2... 61.6 6e-10
ath:AT5G59890 ADF4; ADF4 (ACTIN DEPOLYMERIZING FACTOR 4); acti... 60.8 1e-09
ath:AT3G46010 ADF1; ADF1 (ACTIN DEPOLYMERIZING FACTOR 1); acti... 59.3 3e-09
xla:379258 cfl1-a, MGC54000, cfl1, xac1, xac2; cofilin 1 (non-... 58.2 6e-09
xla:379172 cfl1-b, MGC53097, xac1, xac2; cofilin 1 (non-muscle... 57.4 1e-08
ath:AT3G45990 actin-depolymerizing factor, putative 57.0 1e-08
cpv:cgd5_2800 actin depolymerizing factor ; K05765 cofilin 55.8 3e-08
ath:AT2G16700 ADF5; ADF5 (ACTIN DEPOLYMERIZING FACTOR 5); acti... 55.1 5e-08
pfa:PFE0165w actin-depolymerizing factor, putative 53.9 1e-07
tpv:TP01_0926 actin depolymerizing factor 53.5 2e-07
bbo:BBOV_IV008510 23.m06210; hypothetical protein; K05765 cofilin 53.1 2e-07
ath:AT4G34970 ADF9; ADF9 (ACTIN DEPOLYMERIZING FACTOR 9); acti... 52.4 3e-07
pfa:PF13_0326 actin-depolymerizing factor, putative 51.6 6e-07
hsa:729454 destrin-like 50.8 1e-06
dre:406738 cfl1, cb86, sb:cb86, wu:fb18a04, wu:fk77b03, zgc:56... 49.3 3e-06
xla:379282 dstn, MGC53245; destrin (actin depolymerizing facto... 48.9 4e-06
ath:AT5G59880 ADF3; ADF3 (ACTIN DEPOLYMERIZING FACTOR 3); acti... 41.2 9e-04
dre:100333023 twinfilin-like protein-like 37.0 0.017
dre:553677 MGC112092; zgc:112092; K08870 PTK9 protein tyrosine... 36.6 0.018
cel:C38C3.5 unc-60; UNCoordinated family member (unc-60) 36.6 0.020
sce:YGR080W TWF1; Twf1p 36.2 0.029
dre:432375 twf1b, zgc:92472; twinfilin, actin-binding protein,... 35.8 0.038
dre:100310784 twf2, MGC91817; twinfilin-like protein; K08870 P... 35.4 0.051
dre:325677 twf1a, ptk9, twf1, wu:fd02b03, zgc:65922; twinfilin... 34.3 0.11
mmu:23999 Twf2, A6-related, AU014993, Ptk9l, Ptk9r; twinfilin,... 33.9 0.12
hsa:11344 TWF2, A6RP, A6r, FLJ56277, PTK9L; twinfilin, actin-b... 33.9 0.13
cel:M4.2 puf-4; PUF (Pumilio/FBF) domain-containing family mem... 33.9 0.14
xla:447782 twf2-b, MGC84569, a6r, a6rp, mstp011, ptk9l; twinfi... 33.1 0.20
hsa:5756 TWF1, A6, MGC23788, MGC41876, PTK9; twinfilin, actin-... 33.1 0.20
xla:379995 twf2-a, MGC53423, a6r, a6rp, mstp011, ptk9l, twf2; ... 33.1 0.22
mmu:19230 Twf1, A6, Ptk9, twinfilin; twinfilin, actin-binding ... 32.7 0.26
xla:447307 twf1, MGC81683, ptk9; twinfilin, actin-binding prot... 31.6 0.70
cel:C48B6.4 hypothetical protein 30.8 1.2
> tgo:TGME49_020400 actin depolymerizing factor ; K05765 cofilin
Length=118
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 53/86 (61%), Positives = 63/86 (73%), Gaps = 0/86 (0%)
Query 40 ERNEIVVEKKGTGDASTLTKELPASDCRYAVYDEGQRIHFILWSPDCAPVKPRMIYSSSK 99
E +IVVEK G G+A LPA+DCR+ VYD G +I F+LW PD APVKPRM Y+SSK
Sbjct 32 ENTKIVVEKDGKGNADEFRGALPANDCRFGVYDCGNKIQFVLWCPDNAPVKPRMTYASSK 91
Query 100 DALAKKLEGTVATTLEAHELGDLSVL 125
DAL KKL+G A LEAHE+GDL+ L
Sbjct 92 DALLKKLDGATAVALEAHEMGDLAAL 117
> sce:YLL050C COF1; Cof1p; K05765 cofilin
Length=143
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 61/101 (60%), Gaps = 12/101 (11%)
Query 34 LFCEDGERNEIVVEKKGTGDA-STLTKELPASDCRYAVYD--------EGQR--IHFILW 82
LF + + EIVV++ T + ++LP +DC YA+YD EG+R I F W
Sbjct 29 LFGLNDAKTEIVVKETSTDPSYDAFLEKLPENDCLYAIYDFEYEINGNEGKRSKIVFFTW 88
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLS 123
SPD APV+ +M+Y+SSKDAL + L G V+T ++ + ++S
Sbjct 89 SPDTAPVRSKMVYASSKDALRRALNG-VSTDVQGTDFSEVS 128
> ath:AT4G25590 ADF7; ADF7 (actin depolymerizing factor 7); actin
binding
Length=137
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 61/101 (60%), Gaps = 14/101 (13%)
Query 34 LFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILW 82
+F DG+ ++VVEK G D + T LPA++CRYAV+D + +I FI W
Sbjct 31 IFRIDGQ--QVVVEKLGNPDETYDDFTASLPANECRYAVFDFDFITDENCQKSKIFFIAW 88
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLS 123
SPD + V+ +M+Y+SSKD ++L+G + L+A + ++S
Sbjct 89 SPDSSRVRMKMVYASSKDRFKRELDG-IQVELQATDPSEMS 128
> hsa:1073 CFL2, NEM7; cofilin 2 (muscle); K05765 cofilin
Length=166
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKK------GTGDA-----STLTKELPASDCRYAVYD------- 72
+ LFC ++ +I+VE+ GD ++ K LP +DCRYA+YD
Sbjct 34 KAVLFCLSDDKRQIIVEEAKQILVGDIGDTVEDPYTSFVKLLPLNDCRYALYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KK G
Sbjct 94 SKKEDLVFIFWAPESAPLKSKMIYASSKDAIKKKFTG 130
> mmu:12632 Cfl2; cofilin 2, muscle; K05765 cofilin
Length=166
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKK------GTGDA-----STLTKELPASDCRYAVYD------- 72
+ LFC ++ +I+VE+ GD ++ K LP +DCRYA+YD
Sbjct 34 KAVLFCLSDDKRQIIVEEAKQILVGDIGDTVEDPYTSFVKLLPLNDCRYALYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KK G
Sbjct 94 SKKEDLVFIFWAPESAPLKSKMIYASSKDAIKKKFTG 130
> hsa:1072 CFL1, CFL; cofilin 1 (non-muscle); K05765 cofilin
Length=166
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 54/97 (55%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDA--------STLTKELPASDCRYAVYD------- 72
+ LFC ++ I++E+ GD +T K LP DCRYA+YD
Sbjct 34 KAVLFCLSEDKKNIILEEGKEILVGDVGQTVDDPYATFVKMLPDKDCRYALYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KKL G
Sbjct 94 SKKEDLVFIFWAPESAPLKSKMIYASSKDAIKKKLTG 130
> mmu:12631 Cfl1, AA959946, Cof; cofilin 1, non-muscle; K05765
cofilin
Length=166
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 54/97 (55%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDA--------STLTKELPASDCRYAVYD------- 72
+ LFC ++ I++E+ GD +T K LP DCRYA+YD
Sbjct 34 KAVLFCLSEDKKNIILEEGKEILVGDVGQTVDDPYTTFVKMLPDKDCRYALYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KKL G
Sbjct 94 SKKEDLVFIFWAPENAPLKSKMIYASSKDAIKKKLTG 130
> mmu:100048522 cofilin-1-like; K05765 cofilin
Length=166
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 54/97 (55%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDA--------STLTKELPASDCRYAVYD------- 72
+ LFC ++ I++E+ GD +T K LP DCRYA+YD
Sbjct 34 KAVLFCLSEDKKNIILEEGKEILVGDVGQTVDDPYTTFVKMLPDKDCRYALYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KKL G
Sbjct 94 SKKEDLVFIFWAPENAPLKSKMIYASSKDAIKKKLTG 130
> ath:AT5G52360 ADF10; ADF10 (ACTIN DEPOLYMERIZING FACTOR 10);
actin binding; K05765 cofilin
Length=137
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 61/102 (59%), Gaps = 14/102 (13%)
Query 34 LFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILW 82
+F DG+ ++VVEK G+ + T LP ++CRYAVYD + +I FI W
Sbjct 31 IFRIDGQ--QVVVEKLGSPQENYDDFTNYLPPNECRYAVYDFDFTTAENIQKSKIFFIAW 88
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLSV 124
SPD + V+ +M+Y+SSKD ++L+G + L+A + ++S+
Sbjct 89 SPDSSRVRMKMVYASSKDRFKRELDG-IQVELQATDPSEMSL 129
> xla:494995 cfl2; cofilin 2 (non-muscle); K05765 cofilin
Length=167
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 51/97 (52%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDAS--------TLTKELPASDCRYAVYD------- 72
+ LFC ++ EI+VE+ GD T LP DCRY +YD
Sbjct 34 KAVLFCLSPDKKEIIVEETKQILVGDIGEAVQDPYRTFVNLLPLDDCRYGLYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+PD AP+K +MIY+SSKDA+ KK G
Sbjct 94 SKKEDLVFIFWAPDNAPLKSKMIYASSKDAIKKKFTG 130
> ath:AT3G46000 ADF2; ADF2 (ACTIN DEPOLYMERIZING FACTOR 2); actin
binding
Length=137
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 46/80 (57%), Gaps = 11/80 (13%)
Query 40 ERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILWSPDCAP 88
E +++VEK G + S LPA DCRY +YD + +I FI WSPD A
Sbjct 35 EDKQVIVEKLGEPEQSYDDFAASLPADDCRYCIYDFDFVTAENCQKSKIFFIAWSPDTAK 94
Query 89 VKPRMIYSSSKDALAKKLEG 108
V+ +MIY+SSKD ++L+G
Sbjct 95 VRDKMIYASSKDRFKRELDG 114
> dre:321496 cfl2l, CFL2, wu:fb17d06, wu:fb18d11, wu:fd59f08,
wu:fj34b08; cofilin 2, like; K05765 cofilin
Length=165
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 52/93 (55%), Gaps = 15/93 (16%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDAS----TLTKELPASDCRYAVYD--------EGQ 75
+ +FC ++ I++E+ GD K LP +DCRYA+YD + +
Sbjct 35 KAVMFCLSDDKKHIIMEQGQEILQGDEGDPYLKFVKMLPPNDCRYALYDATYETKETKKE 94
Query 76 RIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ FI W+P+ AP+K +MIY+SSKDA+ KK G
Sbjct 95 DLVFIFWAPESAPLKSKMIYASSKDAIKKKFTG 127
> ath:AT1G01750 ADF11; ADF11 (ACTIN DEPOLYMERIZING FACTOR 11);
actin binding
Length=140
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 60/102 (58%), Gaps = 12/102 (11%)
Query 34 LFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILW 82
+F D + +++++K G + + T+ +P +CRYAVYD + +I FI W
Sbjct 31 VFKIDEKAQQVMIDKLGNPEETYEDFTRSIPEDECRYAVYDYDFTTPENCQKSKIFFIAW 90
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLSV 124
SPD + V+ +M+Y+SSKD ++L+G + L+A + ++S+
Sbjct 91 SPDTSRVRSKMLYASSKDRFKRELDG-IQVELQATDPSEMSL 131
> dre:403001 cfl2, MGC77288, zgc:77288; cofilin 2 (muscle); K05765
cofilin
Length=166
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKK------GTGDA-----STLTKELPASDCRYAVYD------- 72
+ LFC ++ +I+VE+ GD+ + K LP +DCRY +YD
Sbjct 34 KAVLFCLSDDKKKIIVEEGRQILVGDIGDSVDDPYACFVKLLPLNDCRYGLYDATYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + FI W+P+ AP+K +MIY+SSKDA+ KK G
Sbjct 94 SKKEDLVFIFWAPEGAPLKSKMIYASSKDAIKKKFTG 130
> ath:AT4G00680 ADF8; ADF8 (ACTIN DEPOLYMERIZING FACTOR 8); actin
binding
Length=140
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 58/102 (56%), Gaps = 12/102 (11%)
Query 34 LFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILW 82
+F D + ++ +EK G + + T +P +CRYAVYD + +I FI W
Sbjct 31 VFKIDEKAQQVQIEKLGNPEETYDDFTSSIPDDECRYAVYDFDFTTEDNCQKSKIFFIAW 90
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLSV 124
SPD + V+ +M+Y+SSKD +++EG + L+A + ++S+
Sbjct 91 SPDTSRVRSKMLYASSKDRFKREMEG-IQVELQATDPSEMSL 131
> hsa:11034 DSTN, ACTDP, ADF, bA462D18.2; destrin (actin depolymerizing
factor); K10363 destrin (actin-depolymerizing factor)
Length=148
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 51/97 (52%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDASTLTKE--------LPASDCRYAVYD------- 72
+ +FC ++ I+VE+ GD + LP DCRYA+YD
Sbjct 17 KAVIFCLSADKKCIIVEEGKEILVGDVGVTITDPFKHFVGMLPEKDCRYALYDASFETKE 76
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + F LW+P+ AP+K +MIY+SSKDA+ KK +G
Sbjct 77 SRKEELMFFLWAPELAPLKSKMIYASSKDAIKKKFQG 113
> ath:AT2G31200 ADF6; ADF6 (ACTIN DEPOLYMERIZING FACTOR 6); actin
binding
Length=146
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 11/86 (12%)
Query 34 LFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILW 82
+F D + E+VVEK G S LP +DCRYAVYD + +I F W
Sbjct 38 VFKIDESKKEVVVEKTGNPTESYDDFLASLPDNDCRYAVYDFDFVTSENCQKSKIFFFAW 97
Query 83 SPDCAPVKPRMIYSSSKDALAKKLEG 108
SP + ++ +++YS+SKD L+++L+G
Sbjct 98 SPSTSGIRAKVLYSTSKDQLSRELQG 123
> mmu:56431 Dstn, 2610043P17Rik, ADF, AU042046, Dsn, corn1, sid23p;
destrin; K10363 destrin (actin-depolymerizing factor)
Length=165
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 53/97 (54%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGD-ASTLTKE-------LPASDCRYAVYD------- 72
+ +FC ++ IVVE+ GD +T+T LP DCRYA+YD
Sbjct 34 KAVIFCLSADKKCIVVEEGKEILVGDVGATITDPFKHFVGMLPEKDCRYALYDASFETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + F LW+P+ AP+K +MIY+SSKDA+ KK G
Sbjct 94 SRKEELMFFLWAPEQAPLKSKMIYASSKDAIKKKFPG 130
> ath:AT5G59890 ADF4; ADF4 (ACTIN DEPOLYMERIZING FACTOR 4); actin
binding
Length=132
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 57/102 (55%), Gaps = 20/102 (19%)
Query 40 ERNEIVVEKKGTGDASTLTKE-----LPASDCRYAVYD---------EGQRIHFILWSPD 85
++ +++VEK G LT E LPA +CRYA+YD + +I FI W PD
Sbjct 30 KQKQVIVEKVGE---PILTYEDFAASLPADECRYAIYDFDFVTAENCQKSKIFFIAWCPD 86
Query 86 CAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELG--DLSVL 125
A V+ +MIY+SSKD ++L+G + L+A + DL VL
Sbjct 87 VAKVRSKMIYASSKDRFKRELDG-IQVELQATDPTEMDLDVL 127
> ath:AT3G46010 ADF1; ADF1 (ACTIN DEPOLYMERIZING FACTOR 1); actin
binding; K05765 cofilin
Length=150
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 54/96 (56%), Gaps = 12/96 (12%)
Query 40 ERNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD---------EGQRIHFILWSPDCAP 88
++ ++VVEK G + LPA +CRYA+YD + +I FI W PD A
Sbjct 48 KQKQVVVEKVGQPIQTYEEFAACLPADECRYAIYDFDFVTAENCQKSKIFFIAWCPDIAK 107
Query 89 VKPRMIYSSSKDALAKKLEGTVATTLEAHELGDLSV 124
V+ +MIY+SSKD ++L+G + L+A + ++ +
Sbjct 108 VRSKMIYASSKDRFKRELDG-IQVELQATDPTEMDL 142
> xla:379258 cfl1-a, MGC54000, cfl1, xac1, xac2; cofilin 1 (non-muscle);
K05765 cofilin
Length=168
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 8/61 (13%)
Query 56 TLTKELPASDCRYAVYD--------EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLE 107
T K LP +DCRYA+YD + + + F+ W+P+ A +K +MIY+SSKDA+ K+L
Sbjct 70 TFVKMLPRNDCRYALYDALYETKETKKEDLVFVFWAPEEASLKSKMIYASSKDAIKKRLP 129
Query 108 G 108
G
Sbjct 130 G 130
> xla:379172 cfl1-b, MGC53097, xac1, xac2; cofilin 1 (non-muscle);
K05765 cofilin
Length=168
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/97 (32%), Positives = 51/97 (52%), Gaps = 19/97 (19%)
Query 31 QVALFCEDGERNEIVVEKKG---TGDA--------STLTKELPASDCRYAVYD------- 72
+ +FC ++ I++E GD T K LP +DCRYA+YD
Sbjct 34 KAVIFCLSDDKKTIILEPGKEILQGDVGCNVEDPYKTFVKMLPRNDCRYALYDALYETKE 93
Query 73 -EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+ + + F+ W+P+ A +K +MIY+SSKDA+ K+ G
Sbjct 94 TKKEDLVFVFWAPEEASLKSKMIYASSKDAIRKRFTG 130
> ath:AT3G45990 actin-depolymerizing factor, putative
Length=133
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 44/69 (63%), Gaps = 6/69 (8%)
Query 61 LPASDCRYAVYD----EGQR-IHFILWSPDCAPVKPRMIYSSSKDALAKKLEGTVATTLE 115
LPA +CRYA+ D G+R I FI WSP A ++ +MIYSS+KD ++L+G +
Sbjct 58 LPADECRYAILDIEFVPGERKICFIAWSPSTAKMRKKMIYSSTKDRFKRELDG-IQVEFH 116
Query 116 AHELGDLSV 124
A +L D+S+
Sbjct 117 ATDLTDISL 125
> cpv:cgd5_2800 actin depolymerizing factor ; K05765 cofilin
Length=135
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 43/70 (61%), Gaps = 6/70 (8%)
Query 59 KELPASDCRYAVYD----EGQ--RIHFILWSPDCAPVKPRMIYSSSKDALAKKLEGTVAT 112
K +P ++C YA D GQ ++ F++++P+ A VK RM+++SSKD KKLEG
Sbjct 56 KSIPETECFYATIDLPDPNGQTPKLIFLMFTPENAKVKDRMVFASSKDGFVKKLEGVHGK 115
Query 113 TLEAHELGDL 122
L+A E DL
Sbjct 116 LLQASERSDL 125
> ath:AT2G16700 ADF5; ADF5 (ACTIN DEPOLYMERIZING FACTOR 5); actin
binding
Length=143
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 32 VALFCEDGERNEIVVEKKGTGDA-STLTKELPASDCRYAVYD---------EGQRIHFIL 81
+ E+ R V + G G++ L LP DCRYAV+D +I FI
Sbjct 34 IVFKIEEKSRKVTVDKVGGAGESYHDLEDSLPVDDCRYAVFDFDFVTVDNCRKSKIFFIA 93
Query 82 WSPDCAPVKPRMIYSSSKDALAKKLEG 108
WSP+ + ++ +++Y++SKD L + LEG
Sbjct 94 WSPEASKIRAKILYATSKDGLRRVLEG 120
> pfa:PFE0165w actin-depolymerizing factor, putative
Length=122
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 53/89 (59%), Gaps = 11/89 (12%)
Query 43 EIVVEKKGTGDASTLTKELPASD------CRYAVYDEGQRIHFILWSPDCAPVKPRMIYS 96
EI++ KG ++TLT+ + + D C Y V+D +IHF +++ + + + RM Y+
Sbjct 35 EIIIHSKGA--STTLTELVQSIDKNNEIQCAYVVFDAVSKIHFFMYARESSNSRDRMTYA 92
Query 97 SSKDALAKKLEG-TVATTL--EAHELGDL 122
SSK A+ KK+EG V T++ A ++ DL
Sbjct 93 SSKQAILKKIEGVNVLTSVIESAQDVADL 121
> tpv:TP01_0926 actin depolymerizing factor
Length=120
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 44 IVVEKKGTGDASTLTKELPASDCRYAVYDEGQRIHFILWSPDCAPVKPRMIYSSSKDALA 103
+ V+ G GD L LP DC + VYD+GQ + +++P A + R +YS++K +
Sbjct 37 VSVQNDGEGDVEELLTVLPKDDCAFVVYDKGQNLVLFMFAPPGAKTQSRTVYSTTKQTVE 96
Query 104 KKLEG 108
L G
Sbjct 97 NALSG 101
> bbo:BBOV_IV008510 23.m06210; hypothetical protein; K05765 cofilin
Length=120
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 44 IVVEKKGTGDASTLTKELPASDCRYAVYDEGQRIHFILWSPDCAPVKPRMIYSSSKDALA 103
+ V +G+G+ L LP DC + +YD G+ + +++ AP R IYS++K +
Sbjct 36 VTVVNQGSGEVDELYDALPKDDCAFVLYDTGRYVVLFMYASPSAPTNSRTIYSTTKQTVE 95
Query 104 KKLEGT 109
K LEG+
Sbjct 96 KSLEGS 101
> ath:AT4G34970 ADF9; ADF9 (ACTIN DEPOLYMERIZING FACTOR 9); actin
binding
Length=141
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 53/109 (48%), Gaps = 13/109 (11%)
Query 12 WSTSCCRL-FSYLYFSSFILQVALFCEDGERNEIVVEKKGTGDAS--TLTKELPASDCRY 68
W T C+ F + + V E+ R ++ V+K G S L LP DCRY
Sbjct 11 WMTDDCKKSFMEMKWKKVHRYVVYKLEEKSR-KVTVDKVGAAGESYDDLAASLPEDDCRY 69
Query 69 AVYD---------EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
AV+D +I FI WSP+ + ++ +M+Y++SK L + L+G
Sbjct 70 AVFDFDYVTVDNCRMSKIFFITWSPEASRIREKMMYATSKSGLRRVLDG 118
> pfa:PF13_0326 actin-depolymerizing factor, putative
Length=143
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 36/61 (59%), Gaps = 9/61 (14%)
Query 57 LTKELPASDCRYAVYD------EG---QRIHFILWSPDCAPVKPRMIYSSSKDALAKKLE 107
+ L ++CRY + D EG RI+FI WSPD A K +M+Y+SSK+ L +K+
Sbjct 58 IRNNLKTTECRYIIADMPIPTPEGVLRNRIYFIFWSPDLAKSKEKMLYASSKEYLVRKIN 117
Query 108 G 108
G
Sbjct 118 G 118
> hsa:729454 destrin-like
Length=199
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 20/99 (20%)
Query 31 QVALFCEDGERNEIVVEKKG-----------TGDASTLTKELPASDCRYAVYD------E 73
+ +FC ++ I+VE+ TG LP DC YA+YD +
Sbjct 74 KTVIFCLSADKKCIIVEEGKEISAGDIGVTITGPFKHFVGMLPEKDCCYALYDASFETKK 133
Query 74 GQRIHFI---LWSPDCAPVKPRMIYSSSKDALAKKLEGT 109
R+ F+ LW+P+ P+K +MI++S KDA+ KK +
Sbjct 134 SGRVLFVCLFLWAPELPPLKSKMIFTSCKDAIKKKFQAN 172
> dre:406738 cfl1, cb86, sb:cb86, wu:fb18a04, wu:fk77b03, zgc:56501;
cofilin 1 (non-muscle); K05765 cofilin
Length=163
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 35/56 (62%), Gaps = 8/56 (14%)
Query 61 LPASDCRYAVYD--------EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
LP +CRYA+YD + + FI +PD AP++ +M+Y+SSK+AL KL G
Sbjct 72 LPPKECRYALYDCKYTNKESVKEDLVFIFSAPDDAPMRSKMLYASSKNALKAKLPG 127
> xla:379282 dstn, MGC53245; destrin (actin depolymerizing factor);
K05765 cofilin
Length=153
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 11/95 (11%)
Query 38 DGERNEIVVEKKGTGDASTLTKELPASDCRYAVYDEG--------QRIHFILWSPDCAPV 89
D E+ EI+V+ KG TL P C YA+ D Q + F++W+PD A +
Sbjct 42 DKEK-EILVDHKGDF-FQTLKSMFPEKKCCYALIDVNYSTGETLRQDLMFVMWTPDTATI 99
Query 90 KPRMIYSSSKDALAKKLEGTVATTLEAHELGDLSV 124
K +M+++SSK +L + L G V E DL++
Sbjct 100 KQKMLFASSKSSLKQALPG-VQKQWEIQSREDLTL 133
> ath:AT5G59880 ADF3; ADF3 (ACTIN DEPOLYMERIZING FACTOR 3); actin
binding
Length=124
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 32/58 (55%), Gaps = 11/58 (18%)
Query 41 RNEIVVEKKGTGDAS--TLTKELPASDCRYAVYD------EG---QRIHFILWSPDCA 87
+ +++VEK G + L LPA +CRYA++D EG RI F+ WSPD A
Sbjct 38 QKQVIVEKIGEPGQTHEDLAASLPADECRYAIFDFDFVSSEGVPRSRIFFVAWSPDTA 95
> dre:100333023 twinfilin-like protein-like
Length=147
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEGT 109
FI WSPD +PV+ +M+Y++++ L K+ G+
Sbjct 85 FISWSPDQSPVRLKMVYAATRATLKKEFGGS 115
> dre:553677 MGC112092; zgc:112092; K08870 PTK9 protein tyrosine
kinase 9
Length=364
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEGT 109
FI WSPD +PV+ +M+Y++++ L K+ G+
Sbjct 85 FISWSPDQSPVRLKMVYAATRATLKKEFGGS 115
> cel:C38C3.5 unc-60; UNCoordinated family member (unc-60)
Length=212
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 67 RYAVYDEGQRIHFILWS--PDCAPVKPRMIYSSSKDALAKKLEGTVATTLEAHELGDL 122
++ V DE + H L + PD APV+ RM+Y+SS AL L ++A E+ DL
Sbjct 140 QFQVSDESEMSHKELLNNCPDNAPVRRRMLYASSVRALKASLGLESLFQVQASEMSDL 197
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 13/56 (23%)
Query 64 SDCRYAVYD-------------EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKL 106
+DCRYAV+D + +I F+ PD A +K +M+Y+SS A+ L
Sbjct 78 TDCRYAVFDFKFTCSRVGAGTSKMDKIIFLQICPDGASIKKKMVYASSAAAIKTSL 133
> sce:YGR080W TWF1; Twf1p
Length=332
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 31/49 (63%), Gaps = 5/49 (10%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEGTVATT-----LEAHELGDL 122
F+ + PD +PV+ RM+Y+S+K+ LA+++ +T +A +L DL
Sbjct 77 FVSFIPDGSPVRSRMLYASTKNTLARQVGSNSLSTEQPLITDAQDLVDL 125
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query 68 YAVYDEGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEGTVATTL-EAHELGDL 122
Y ++ +G FI P + VK RMIY+S+K+ L+ + E+GD
Sbjct 232 YTIFRQGDSSFFIYSCPSGSKVKDRMIYASNKNGFINYLKNDQKIAFSKVVEIGDF 287
> dre:432375 twf1b, zgc:92472; twinfilin, actin-binding protein,
homolog 1b; K08870 PTK9 protein tyrosine kinase 9
Length=349
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
F+ WSPD +PV+ +M+Y++++ L K+ G
Sbjct 85 FLAWSPDHSPVRQKMLYAATRATLKKEFGG 114
> dre:100310784 twf2, MGC91817; twinfilin-like protein; K08870
PTK9 protein tyrosine kinase 9
Length=347
Score = 35.4 bits (80), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
FI WSPD +PV+ +M+Y++++ + K+ G
Sbjct 83 FISWSPDQSPVRQKMLYAATRATVKKEFGG 112
> dre:325677 twf1a, ptk9, twf1, wu:fd02b03, zgc:65922; twinfilin,
actin-binding protein, homolog 1a; K08870 PTK9 protein tyrosine
kinase 9
Length=350
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKL 106
FI WSPD +PV+ +M+Y++++ + K+
Sbjct 85 FIAWSPDHSPVRHKMLYAATRATIKKEF 112
> mmu:23999 Twf2, A6-related, AU014993, Ptk9l, Ptk9r; twinfilin,
actin-binding protein, homolog 2 (Drosophila); K08870 PTK9
protein tyrosine kinase 9
Length=349
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
F+ WSPD +PV+ +M+Y++++ + K+ G
Sbjct 85 FLAWSPDNSPVRLKMLYAATRATVKKEFGG 114
> hsa:11344 TWF2, A6RP, A6r, FLJ56277, PTK9L; twinfilin, actin-binding
protein, homolog 2 (Drosophila); K08870 PTK9 protein
tyrosine kinase 9
Length=349
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
F+ WSPD +PV+ +M+Y++++ + K+ G
Sbjct 85 FLAWSPDNSPVRLKMLYAATRATVKKEFGG 114
> cel:M4.2 puf-4; PUF (Pumilio/FBF) domain-containing family member
(puf-4)
Length=420
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 25 FSSFILQVALFCEDGERNEIVVEKKGTGDASTLTKELPASDCRYAVYDE 73
F +F +Q L C D E +I+VE AS L EL D +A D+
Sbjct 207 FGNFFVQRVLECSDAEEQKIIVEYLAIVLASRLASELRGVDLTHACIDQ 255
> xla:447782 twf2-b, MGC84569, a6r, a6rp, mstp011, ptk9l; twinfilin,
actin-binding protein, homolog 2; K08870 PTK9 protein
tyrosine kinase 9
Length=349
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
F+ WSPD +PV+ +M+Y++++ + K+ G
Sbjct 85 FLSWSPDHSPVRLKMLYAATRATVKKEFGG 114
> hsa:5756 TWF1, A6, MGC23788, MGC41876, PTK9; twinfilin, actin-binding
protein, homolog 1 (Drosophila); K08870 PTK9 protein
tyrosine kinase 9
Length=384
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 73 EGQRIHFILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
+G FI WSPD + V+ +M+Y++++ L K+ G
Sbjct 113 QGYEWIFIAWSPDHSHVRQKMLYAATRATLKKEFGG 148
> xla:379995 twf2-a, MGC53423, a6r, a6rp, mstp011, ptk9l, twf2;
twinfilin, actin-binding protein, homolog 2; K08870 PTK9 protein
tyrosine kinase 9
Length=349
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
F+ WSPD +PV+ +M+Y++++ + K+ G
Sbjct 85 FLSWSPDHSPVRLKMLYAATRATVKKEFGG 114
> mmu:19230 Twf1, A6, Ptk9, twinfilin; twinfilin, actin-binding
protein, homolog 1 (Drosophila); K08870 PTK9 protein tyrosine
kinase 9
Length=350
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
FI WSPD + V+ +M+Y++++ L K+ G
Sbjct 85 FIAWSPDHSHVRQKMLYAATRATLKKEFGG 114
> xla:447307 twf1, MGC81683, ptk9; twinfilin, actin-binding protein,
homolog 1; K08870 PTK9 protein tyrosine kinase 9
Length=350
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 79 FILWSPDCAPVKPRMIYSSSKDALAKKLEG 108
FI WSPD + V+ +M+Y++++ + K+ G
Sbjct 85 FIAWSPDYSHVRQKMLYAATRATVKKEFGG 114
> cel:C48B6.4 hypothetical protein
Length=623
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 40 ERNEIVVEKKGTGDASTLTKELPASDCRYAVYDEGQRIHFI 80
ERN ++EKKG+ +T+ + A++ R A+Y++ + F+
Sbjct 395 ERNSYLIEKKGSSVPDFVTQPIQAANARKALYEKSCYLGFL 435
Lambda K H
0.320 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40