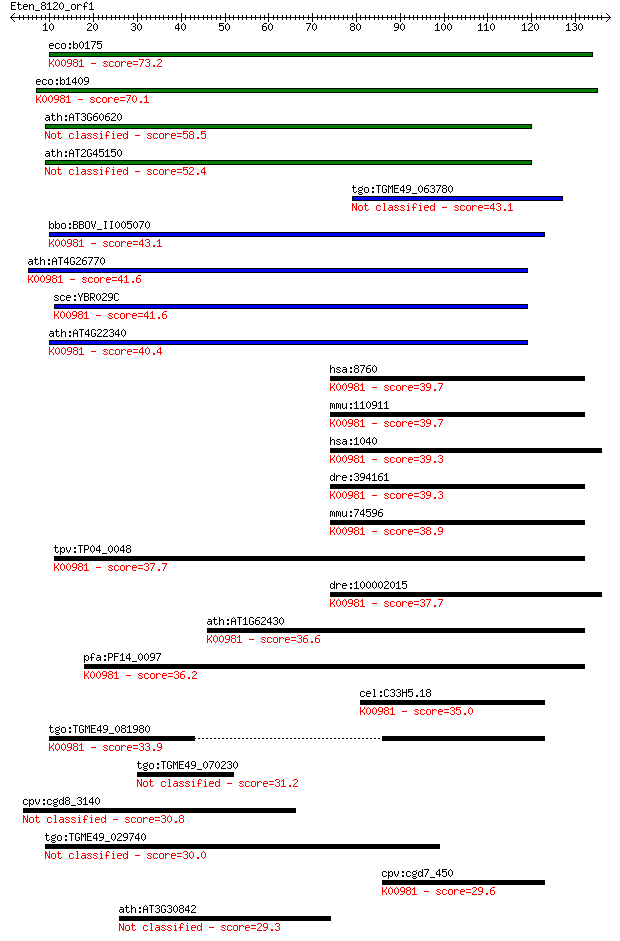
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8120_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
eco:b0175 cdsA, ECK0174, JW5810; CDP-diglyceride synthase (EC:... 73.2 2e-13
eco:b1409 ynbB, ECK1402, JW1406; predicted CDP-diglyceride syn... 70.1 2e-12
ath:AT3G60620 phosphatidate cytidylyltransferase family protein 58.5 6e-09
ath:AT2G45150 phosphatidate cytidylyltransferase family protei... 52.4 4e-07
tgo:TGME49_063780 phosphatidate cytidylyltransferase, putative... 43.1 3e-04
bbo:BBOV_II005070 18.m06422; cytidine diphosphate-diacylglycer... 43.1 3e-04
ath:AT4G26770 phosphatidate cytidylyltransferase/ transferase,... 41.6 7e-04
sce:YBR029C CDS1, CDG1; Cds1p (EC:2.7.7.41); K00981 phosphatid... 41.6 8e-04
ath:AT4G22340 phosphatidate cytidylyltransferase, putative / C... 40.4 0.002
hsa:8760 CDS2, FLJ38111; CDP-diacylglycerol synthase (phosphat... 39.7 0.003
mmu:110911 Cds2, 5730450N06Rik, 5730460C18Rik, AI854580, D2Wsu... 39.7 0.003
hsa:1040 CDS1, CDS; CDP-diacylglycerol synthase (phosphatidate... 39.3 0.004
dre:394161 cds2, MGC66134, zgc:66134; CDP-diacylglycerol synth... 39.3 0.004
mmu:74596 Cds1, 4833409J18Rik, AI314024, AW125888; CDP-diacylg... 38.9 0.004
tpv:TP04_0048 phosphatidate cytidylyltransferase (EC:2.7.7.41)... 37.7 0.011
dre:100002015 CDP-diacylglycerol synthase 1-like; K00981 phosp... 37.7 0.012
ath:AT1G62430 ATCDS1; ATCDS1; phosphatidate cytidylyltransfera... 36.6 0.021
pfa:PF14_0097 CDS; cytidine diphosphate-diacylglycerol synthas... 36.2 0.027
cel:C33H5.18 hypothetical protein; K00981 phosphatidate cytidy... 35.0 0.070
tgo:TGME49_081980 cytidine diphosphate-diacylglycerol synthase... 33.9 0.18
tgo:TGME49_070230 hypothetical protein 31.2 1.1
cpv:cgd8_3140 MRP like MinD family ATpase of the SIMIBI class ... 30.8 1.4
tgo:TGME49_029740 hypothetical protein 30.0 2.3
cpv:cgd7_450 cytidine diphosphate-diacylglycerol synthase; int... 29.6 2.6
ath:AT3G30842 PDR10; PDR10 (PLEIOTROPIC DRUG RESISTANCE 10); A... 29.3 3.9
> eco:b0175 cdsA, ECK0174, JW5810; CDP-diglyceride synthase (EC:2.7.7.41);
K00981 phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=285
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 46/124 (37%), Positives = 66/124 (53%), Gaps = 7/124 (5%)
Query 10 GDSGALFCGSAFGKRKIIEKLSPNKTVAGVLGSLSWSWLAGWLLWLLSLKAPYLGLRVLS 69
DSGA G FGK K+ K+SP KT G +G L+ + + W + L V
Sbjct 164 ADSGAYMFGKLFGKHKLAPKVSPGKTWQGFIGGLATAAVISWG------YGMWANLDVAP 217
Query 70 LAEYLCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVF 129
+ +C + + +++ GDL ES FKR AG KDS L HGG+LDR+DS+T +P
Sbjct 218 VTLLIC-SIVAALASVLGDLTESMFKREAGIKDSGHLIPGHGGILDRIDSLTAAVPVFAC 276
Query 130 FMVL 133
++L
Sbjct 277 LLLL 280
> eco:b1409 ynbB, ECK1402, JW1406; predicted CDP-diglyceride synthase;
K00981 phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=298
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 68/128 (53%), Gaps = 10/128 (7%)
Query 7 SAAGDSGALFCGSAFGKRKIIEKLSPNKTVAGVLGSLSWSWLAGWLLWLLSLKAPYLGLR 66
+ + D G + G+RK++ K+SP KT+ G++G + +A ++ P L
Sbjct 177 TESNDIAQYLWGKSCGRRKVVPKVSPGKTLEGLMGGVITIMIASLII------GPLL--T 228
Query 67 VLSLAEYLCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPF 126
L+ + L LI ISGF GD+V S KR G KDS L HGG+LDR+DS+ P
Sbjct 229 PLNTLQALLAGLLIGISGFCGDVVMSAIKRDIGVKDSGKLLPGHGGLLDRIDSLIFTAP- 287
Query 127 LVFFMVLR 134
VFF +R
Sbjct 288 -VFFYFIR 294
> ath:AT3G60620 phosphatidate cytidylyltransferase family protein
Length=399
Score = 58.5 bits (140), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 46/115 (40%), Positives = 58/115 (50%), Gaps = 16/115 (13%)
Query 9 AGDSGALFCGSAFGKRKIIEKLSPNKT----VAGVLGSLSWSWLAGWLLWLLSLKAPYLG 64
A D+ A G AFG+ +I +SP KT AG++G +S + +L SL P
Sbjct 280 ASDTFAFLGGKAFGRTPLIS-ISPKKTWEGAFAGLVGCISIT-----ILLSKSLSWPQ-- 331
Query 65 LRVLSLAEYLCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDS 119
SL + F L FGDL ES KR AG KDS L HGG+LDR+DS
Sbjct 332 ----SLVSTIAFGVLNFFGSVFGDLTESMIKRDAGVKDSGSLIPGHGGILDRVDS 382
> ath:AT2G45150 phosphatidate cytidylyltransferase family protein
(EC:2.7.7.41)
Length=430
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 57/116 (49%), Gaps = 18/116 (15%)
Query 9 AGDSGALFCGSAFGKRKIIEKLSPNKT----VAGVLGSLSWS-WLAGWLLWLLSLKAPYL 63
A D+ A G FG R + +SP KT + G++G ++ + L+ +L W SL +
Sbjct 307 ATDTFAFLGGKTFG-RTPLTSISPKKTWEGTIVGLVGCIAITILLSKYLSWPQSLFSS-- 363
Query 64 GLRVLSLAEYLCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDS 119
+ F L FGDL ES KR AG KDS L HGG+LDR+DS
Sbjct 364 ----------VAFGFLNFFGSVFGDLTESMIKRDAGVKDSGSLIPGHGGILDRVDS 409
> tgo:TGME49_063780 phosphatidate cytidylyltransferase, putative
(EC:2.7.7.41)
Length=551
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 79 LISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPF 126
L+S+ G GDL S KR AG KDS L HGG LDR DS L P
Sbjct 468 LLSLVGVLGDLTASLVKRDAGVKDSGTLLPGHGGWLDRTDSYLLAAPL 515
> bbo:BBOV_II005070 18.m06422; cytidine diphosphate-diacylglycerol
synthase (EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=438
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 41/160 (25%), Positives = 60/160 (37%), Gaps = 51/160 (31%)
Query 10 GDSGALFCGSAFGKRKIIEKLSPNKTVAGVLGSLSWSWLAGWLL---------------- 53
D A F G FGK +I LSPNKT+ G +G+ + + ++
Sbjct 236 NDVMAYFFGKMFGKTPLIS-LSPNKTLEGFIGAFISTTMIVAIMSPIILRFQPMICSPNT 294
Query 54 -------WLLSLKAPYLGLRVLSLAEYL------------------CFATLISISGF--- 85
W+ + P R+ L Y+ CF ++ ++ F
Sbjct 295 FVFTPFAWMRATCEPE---RIYKLTTYVLPTWLSNIVGINEILYRPCFVHIMILTLFASL 351
Query 86 ---FGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTL 122
FG + SGFKR KD S HGG++DR D L
Sbjct 352 FAPFGGFLASGFKRALKIKDFSDTIPGHGGMMDRFDCHIL 391
> ath:AT4G26770 phosphatidate cytidylyltransferase/ transferase,
transferring phosphorus-containing groups (EC:2.7.7.41);
K00981 phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=471
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 54/154 (35%), Gaps = 42/154 (27%)
Query 5 AISAAGDSGALFCGSAFGKRKIIEKLSPNKT-----------------VAGVLGSLSW-- 45
A+ A D A F G FGK +I KLSP KT A VLG W
Sbjct 261 ALIAMNDVAAYFFGFYFGKTPLI-KLSPKKTWEGFIGASVATIISAFIFANVLGQFQWLT 319
Query 46 ----SWLAGWL----------------LWLLSLKAPYLGLRVLSLAEY-LCFATLISISG 84
GWL W+ +P+ G+ L + + SI
Sbjct 320 CPRKDLSTGWLHCDPGPLFRPEYYPFPSWITPF-SPWKGISTLPVQWHAFSLGLFASIMA 378
Query 85 FFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLD 118
FG SGFKR KD HGG DR+D
Sbjct 379 PFGGFFASGFKRAFKIKDFGDSIPGHGGFTDRMD 412
> sce:YBR029C CDS1, CDG1; Cds1p (EC:2.7.7.41); K00981 phosphatidate
cytidylyltransferase [EC:2.7.7.41]
Length=457
Score = 41.6 bits (96), Expect = 8e-04, Method: Composition-based stats.
Identities = 43/150 (28%), Positives = 55/150 (36%), Gaps = 43/150 (28%)
Query 11 DSGALFCGSAFGKRKIIEKLSPNKTVAGVLGSLSWSWLAGWLLW-LLS----LKAPYLGL 65
D A CG FGK K+IE +SP KT+ G LG+ ++ LA +L +LS L P L
Sbjct 229 DIFAYLCGITFGKTKLIE-ISPKKTLEGFLGAWFFTALASIILTRILSPYTYLTCPVEDL 287
Query 66 RVLSLAEYLC-------------------------------------FATLISISGFFGD 88
+ C AT S+ FG
Sbjct 288 HTNFFSNLTCELNPVFLPQVYRLPPIFFDKVQINSITVKPIYFHALNLATFASLFAPFGG 347
Query 89 LVESGFKRLAGAKDSSGLFGPHGGVLDRLD 118
SG KR KD HGG+ DR+D
Sbjct 348 FFASGLKRTFKVKDFGHSIPGHGGITDRVD 377
> ath:AT4G22340 phosphatidate cytidylyltransferase, putative /
CDP-diglyceride synthetase, putative (EC:2.7.7.41); K00981
phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=447
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 52/148 (35%), Gaps = 40/148 (27%)
Query 10 GDSGALFCGSAFGKRKIIEKLSPNKTVAGVLGSLSWSWLAGWLL---------------- 53
D A CG FG+ +I KLSP KT G +G+ + ++ +LL
Sbjct 243 NDIFAYICGFFFGRTPLI-KLSPKKTWEGFIGASITTVISAFLLANIMGRFLWLTCPRED 301
Query 54 ----WLLSLKAPYLGLRVLSLA-------------------EYLCFATLISISGFFGDLV 90
WLL P +L LC SI FG
Sbjct 302 LSTGWLLCDPGPLFKQETHALPGWISDWLPWKEISILPVQWHALCLGLFASIIAPFGGFF 361
Query 91 ESGFKRLAGAKDSSGLFGPHGGVLDRLD 118
SGFKR KD HGG+ DR+D
Sbjct 362 ASGFKRAFKIKDFGDSIPGHGGITDRMD 389
> hsa:8760 CDS2, FLJ38111; CDP-diacylglycerol synthase (phosphatidate
cytidylyltransferase) 2 (EC:2.7.7.41); K00981 phosphatidate
cytidylyltransferase [EC:2.7.7.41]
Length=445
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFM 131
+ +T S+ G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 340 IALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYI 397
> mmu:110911 Cds2, 5730450N06Rik, 5730460C18Rik, AI854580, D2Wsu127e;
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase)
2 (EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=444
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFM 131
+ +T S+ G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 339 IALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYI 396
> hsa:1040 CDS1, CDS; CDP-diacylglycerol synthase (phosphatidate
cytidylyltransferase) 1 (EC:2.7.7.41); K00981 phosphatidate
cytidylyltransferase [EC:2.7.7.41]
Length=461
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 32/64 (50%), Gaps = 2/64 (3%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFMV- 132
+ +T S+ G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 357 IALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVHVYITS 416
Query 133 -LRG 135
+RG
Sbjct 417 FIRG 420
> dre:394161 cds2, MGC66134, zgc:66134; CDP-diacylglycerol synthase
(phosphatidate cytidylyltransferase) 2 (EC:2.7.7.41);
K00981 phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=444
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFM 131
+ ++ SI G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 339 IALSSFASIMGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYI 396
> mmu:74596 Cds1, 4833409J18Rik, AI314024, AW125888; CDP-diacylglycerol
synthase 1 (EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=461
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFM 131
+ +T S+ G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 357 IALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVHVYI 414
> tpv:TP04_0048 phosphatidate cytidylyltransferase (EC:2.7.7.41);
K00981 phosphatidate cytidylyltransferase [EC:2.7.7.41]
Length=457
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 55/128 (42%), Gaps = 17/128 (13%)
Query 11 DSGALFC-GSAFGKRKIIEKLSPNKTVAGVLGSLSWSW--LAGWLLWLLSLKAPY----L 63
+ L C + F R + S N + GV W LA LL L PY L
Sbjct 304 NYKPLLCPTNHFNLRPFVWLHSTNCKLPGVYQIKKWELPELAAKLLKRTHL--PYSEFVL 361
Query 64 GLRVLSLAEYLCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLP 123
+ VLSL FA+L + FG + SGFKR KD S + HGG+ DR D L
Sbjct 362 HMLVLSL-----FASLFAP---FGGFLASGFKRALKVKDFSNVIPGHGGITDRFDCHILM 413
Query 124 IPFLVFFM 131
F F++
Sbjct 414 GGFTYFYL 421
> dre:100002015 CDP-diacylglycerol synthase 1-like; K00981 phosphatidate
cytidylyltransferase [EC:2.7.7.41]
Length=452
Score = 37.7 bits (86), Expect = 0.012, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 2/64 (3%)
Query 74 LCFATLISISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFMV- 132
+ ++ S+ G FG SGFKR KD + HGG++DR D L F+ ++
Sbjct 346 IALSSFASLIGPFGGFFASGFKRAFKIKDFADTIPGHGGIMDRFDCQYLMATFVHVYITS 405
Query 133 -LRG 135
+RG
Sbjct 406 FIRG 409
> ath:AT1G62430 ATCDS1; ATCDS1; phosphatidate cytidylyltransferase
(EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=421
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 36/87 (41%), Gaps = 9/87 (10%)
Query 46 SWLAGWLLWLLSLKAPYLGLRVLSLAEY-LCFATLISISGFFGDLVESGFKRLAGAKDSS 104
+W+ W P+ + +L + + LC SI FG SGFKR KD
Sbjct 300 AWIPEWF--------PWKEMTILPVQWHALCLGLFASIIAPFGGFFASGFKRAFKIKDFG 351
Query 105 GLFGPHGGVLDRLDSMTLPIPFLVFFM 131
HGG+ DR+D + F ++
Sbjct 352 DSIPGHGGITDRMDCQMVMAVFAYIYL 378
> pfa:PF14_0097 CDS; cytidine diphosphate-diacylglycerol synthase
(EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=667
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 42/162 (25%), Positives = 54/162 (33%), Gaps = 52/162 (32%)
Query 18 GSAFGKRKIIEKLSPNKTVAGVLGS----LSWSWLAGWLL-------------------- 53
G FGK ++I+ LSP KTV G +GS + W L + L
Sbjct 432 GILFGKTRLIQ-LSPKKTVEGYVGSSIITIVWGILITYFLQRYKFFICPQKYITFQPFVS 490
Query 54 W------------------------LLSLKAPYLGLRVLSLAEYLCFATLISISGFFGDL 89
W +LS+K Y + FA ++ FG
Sbjct 491 WNYIDCDINPIFQQKVYEVPKQISQILSIKNIYYSKMIFHGLMLSLFAAFLAP---FGGF 547
Query 90 VESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTLPIPFLVFFM 131
SGFKR KD HGGV DR D F +M
Sbjct 548 FASGFKRALKIKDFGKSIPGHGGVTDRFDCQIFIGMFTYIYM 589
> cel:C33H5.18 hypothetical protein; K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=455
Score = 35.0 bits (79), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 81 SISGFFGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTL 122
SI G FG SGFKR KD + HGG++DR D L
Sbjct 364 SILGPFGGFFASGFKRAFKIKDFGDVIPGHGGLMDRFDCQLL 405
> tgo:TGME49_081980 cytidine diphosphate-diacylglycerol synthase,
putative (EC:2.7.7.41); K00981 phosphatidate cytidylyltransferase
[EC:2.7.7.41]
Length=1068
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 86 FGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTL 122
FG SGFKR A KD + HGGV DR D L
Sbjct 849 FGGFFASGFKRAARIKDFGEIIPGHGGVTDRFDCQIL 885
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 10 GDSGALFCGSAFGKRKIIEKLSPNKTVAGVLGS 42
D A CG FG+ ++I +LSP KTV G +G+
Sbjct 592 NDVSAYICGMLFGRTRLI-RLSPKKTVEGFVGA 623
> tgo:TGME49_070230 hypothetical protein
Length=1815
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 30 LSPNKTVAGVLGSLSWSWLAGW 51
L P + V G+ GS S SWLAGW
Sbjct 463 LHPAQEVPGLRGSASPSWLAGW 484
> cpv:cgd8_3140 MRP like MinD family ATpase of the SIMIBI class
of P-loop GTpases
Length=355
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 33/68 (48%), Gaps = 6/68 (8%)
Query 4 IAISAAGDSGALFCGSAFGKRK---IIEKLSPNKTVAGVL---GSLSWSWLAGWLLWLLS 57
IA S AG AL C S K+K IE LS K + VL G + S ++ + W LS
Sbjct 76 IADSCAGCPNALICASGQAKKKPTENIENLSKIKNIILVLSGKGGVGKSTISSQISWCLS 135
Query 58 LKAPYLGL 65
K +GL
Sbjct 136 SKKFNVGL 143
> tgo:TGME49_029740 hypothetical protein
Length=376
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 39/94 (41%), Gaps = 13/94 (13%)
Query 9 AGDSGALFCGSAFGK----RKIIEKLSPNKTVAGVLGSLSWSWLAGWLLWLLSLKAPYLG 64
GD+ + C S +I E PN+ G L S+ W + L+L + YL
Sbjct 202 QGDTCYVLCRSERDAGVVLARIQETAVPNRVHYGQLFGCSFLWASRSALFLCDSQLDYLP 261
Query 65 LRVLSLAEYLCFATLISISGFFGDLVESGFKRLA 98
R + + F T G+ GD+ E FK LA
Sbjct 262 AR----SPFQVFTT-----GWEGDVSEEEFKNLA 286
> cpv:cgd7_450 cytidine diphosphate-diacylglycerol synthase; integral
membrane protein with 7 or more ; K00981 phosphatidate
cytidylyltransferase [EC:2.7.7.41]
Length=450
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 16/37 (43%), Gaps = 0/37 (0%)
Query 86 FGDLVESGFKRLAGAKDSSGLFGPHGGVLDRLDSMTL 122
FG SG KR KD HGG+ DR D L
Sbjct 362 FGGFFASGLKRALRIKDFGSAIPGHGGITDRFDCQIL 398
> ath:AT3G30842 PDR10; PDR10 (PLEIOTROPIC DRUG RESISTANCE 10);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding
Length=1406
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 30/61 (49%), Gaps = 13/61 (21%)
Query 26 IIEKLSPNKTVAGVLG---SLSWSWLAG----------WLLWLLSLKAPYLGLRVLSLAE 72
++ +SPN+ +A +L S SW+ +G WL W + + GL L++A+
Sbjct 1288 MVISVSPNQEIASILNGVISTSWNVFSGFTIPRPRMHVWLRWFTYVCPGWWGLYGLTIAQ 1347
Query 73 Y 73
Y
Sbjct 1348 Y 1348
Lambda K H
0.326 0.144 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40