bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8119_orf2
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
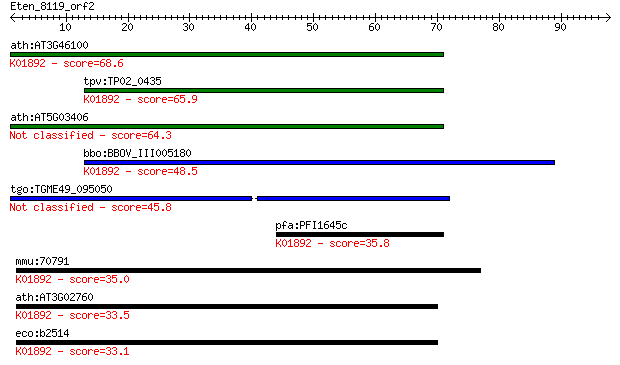
ath:AT3G46100 ATHRS1; ATHRS1 (HISTIDYL-TRNA SYNTHETASE 1); his... 68.6 5e-12
tpv:TP02_0435 histidyl-tRNA synthetase; K01892 histidyl-tRNA s... 65.9 3e-11
ath:AT5G03406 ATP binding / aminoacyl-tRNA ligase/ histidine-t... 64.3 8e-11
bbo:BBOV_III005180 17.m07463; hypothetical protein; K01892 his... 48.5 6e-06
tgo:TGME49_095050 histidyl-tRNA synthetase, putative (EC:6.1.1... 45.8 3e-05
pfa:PFI1645c histidyl-tRNA synthetase, putative; K01892 histid... 35.8 0.036
mmu:70791 Hars2, 4631412B19Rik, AI593507, HARSR, HO3, Harsl; h... 35.0 0.055
ath:AT3G02760 ATP binding / aminoacyl-tRNA ligase/ histidine-t... 33.5 0.19
eco:b2514 hisS, ECK2510, JW2498; histidyl tRNA synthetase (EC:... 33.1 0.25
> ath:AT3G46100 ATHRS1; ATHRS1 (HISTIDYL-TRNA SYNTHETASE 1); histidine-tRNA
ligase (EC:6.1.1.21); K01892 histidyl-tRNA synthetase
[EC:6.1.1.21]
Length=486
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 31/70 (44%), Positives = 46/70 (65%), Gaps = 2/70 (2%)
Query 1 HIYRLHVSGPDPLCLRPEITPSLARMLQREQQQQPSGIARRWYSIERVWRHERPGCGRRR 60
+Y G + LRPE+TPSLAR++ Q+ + + +W++I + WR+ER GRRR
Sbjct 127 QLYCFEDRGNRRVALRPELTPSLARLVI--QKGKSVSLPLKWFAIGQCWRYERMTRGRRR 184
Query 61 EHFQWNLDII 70
EH+QWN+DII
Sbjct 185 EHYQWNMDII 194
> tpv:TP02_0435 histidyl-tRNA synthetase; K01892 histidyl-tRNA
synthetase [EC:6.1.1.21]
Length=508
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 29/60 (48%), Positives = 36/60 (60%), Gaps = 2/60 (3%)
Query 13 LCLRPEITPSLARMLQR--EQQQQPSGIARRWYSIERVWRHERPGCGRRREHFQWNLDII 70
L LR +ITP ML ++ PS +WY+I WR+ERPG GRRR H QWN DI+
Sbjct 120 LSLRGDITPQFMNMLTSYYSNKRVPSHNILKWYTIADCWRYERPGLGRRRNHLQWNADIV 179
> ath:AT5G03406 ATP binding / aminoacyl-tRNA ligase/ histidine-tRNA
ligase/ nucleotide binding
Length=263
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 46/70 (65%), Gaps = 2/70 (2%)
Query 1 HIYRLHVSGPDPLCLRPEITPSLARMLQREQQQQPSGIARRWYSIERVWRHERPGCGRRR 60
+Y G + LRPE+TPSLAR++ Q+ + + +W+++ + WR+ER GRRR
Sbjct 127 QLYCFEDRGNRRVALRPELTPSLARLVI--QKGKSVSLPLKWFAVGQCWRYERMTRGRRR 184
Query 61 EHFQWNLDII 70
EH+QWN+DII
Sbjct 185 EHYQWNMDII 194
> bbo:BBOV_III005180 17.m07463; hypothetical protein; K01892 histidyl-tRNA
synthetase [EC:6.1.1.21]
Length=548
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 41/85 (48%), Gaps = 21/85 (24%)
Query 13 LCLRPEITPSLARMLQREQQQQP---------SGIARRWYSIERVWRHERPGCGRRREHF 63
L LR ++TP ML +E + P + +W+++ WR+ERPG RRR H
Sbjct 163 LALRGDVTPQFMAML-KEHWELPIKHNLNALETKQITKWFTMADCWRYERPGHCRRRNHI 221
Query 64 QWNLDIILPLFPETNPSGGIPSEAA 88
QW DI+ GIP+E A
Sbjct 222 QWTCDIV-----------GIPNEDA 235
> tgo:TGME49_095050 histidyl-tRNA synthetase, putative (EC:6.1.1.21)
Length=1321
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/31 (64%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 41 RWYSIERVWRHERPGCGRRREHFQWNLDIIL 71
R SI WR+ERPG RRR+H+QWNLDI+L
Sbjct 404 RLTSIGDCWRYERPGRCRRRQHWQWNLDIVL 434
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 26/42 (61%), Gaps = 3/42 (7%)
Query 1 HIYRLHVSGPDPLCLRPEITPSLARMLQR---EQQQQPSGIA 39
H+Y PL LRPE+TPSL R+L R + Q++ SG++
Sbjct 253 HLYLFRDQSERPLALRPELTPSLCRLLLRLYAKPQRRVSGVS 294
> pfa:PFI1645c histidyl-tRNA synthetase, putative; K01892 histidyl-tRNA
synthetase [EC:6.1.1.21]
Length=642
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 44 SIERVWRHERPGCGRRREHFQWNLDII 70
+I + +R+E+ R+REH+QWNLDI+
Sbjct 268 TIGQCFRYEQTSRLRKREHYQWNLDIL 294
> mmu:70791 Hars2, 4631412B19Rik, AI593507, HARSR, HO3, Harsl;
histidyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.21);
K01892 histidyl-tRNA synthetase [EC:6.1.1.21]
Length=505
Score = 35.0 bits (79), Expect = 0.055, Method: Composition-based stats.
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 10/80 (12%)
Query 2 IYRLHVSGPDPLCLRPEITPSLARMLQREQQQQPSGIARRWYSIERVWRHERPGC--GRR 59
+Y L G + L LR ++T AR L + ++ + Y + +VWR E P GR
Sbjct 114 MYDLKDQGGELLSLRYDLTVPFARYLAMNKLKK-----MKRYQVGKVWRRESPAIAQGRY 168
Query 60 REHFQWNLDI---ILPLFPE 76
RE Q + DI P+ P+
Sbjct 169 REFCQCDFDIAGEFDPMIPD 188
> ath:AT3G02760 ATP binding / aminoacyl-tRNA ligase/ histidine-tRNA
ligase/ nucleotide binding (EC:6.1.1.21); K01892 histidyl-tRNA
synthetase [EC:6.1.1.21]
Length=883
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query 2 IYRLHVSGPDPLCLRPEITPSLARMLQREQQQQPSGIAR-RWYSIERVWRHERPGCGRRR 60
+Y + G + LR ++T AR + +GI + Y I +V+R + P GR R
Sbjct 504 VYDIADQGGELCSLRYDLTVPFARYVAM------NGITSFKRYQIAKVYRRDNPSKGRYR 557
Query 61 EHFQWNLDI 69
E +Q + DI
Sbjct 558 EFYQCDFDI 566
> eco:b2514 hisS, ECK2510, JW2498; histidyl tRNA synthetase (EC:6.1.1.21);
K01892 histidyl-tRNA synthetase [EC:6.1.1.21]
Length=424
Score = 33.1 bits (74), Expect = 0.25, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 32/73 (43%), Gaps = 12/73 (16%)
Query 2 IYRLHVSGPDPLCLRPEITPSLARM-----LQREQQQQPSGIARRWYSIERVWRHERPGC 56
+Y D L LRPE T R L Q+Q R WY I ++RHERP
Sbjct 67 MYTFEDRNGDSLTLRPEGTAGCVRAGIEHGLLYNQEQ------RLWY-IGPMFRHERPQK 119
Query 57 GRRREHFQWNLDI 69
GR R+ Q ++
Sbjct 120 GRYRQFHQLGCEV 132
Lambda K H
0.321 0.138 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40