bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8117_orf1
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
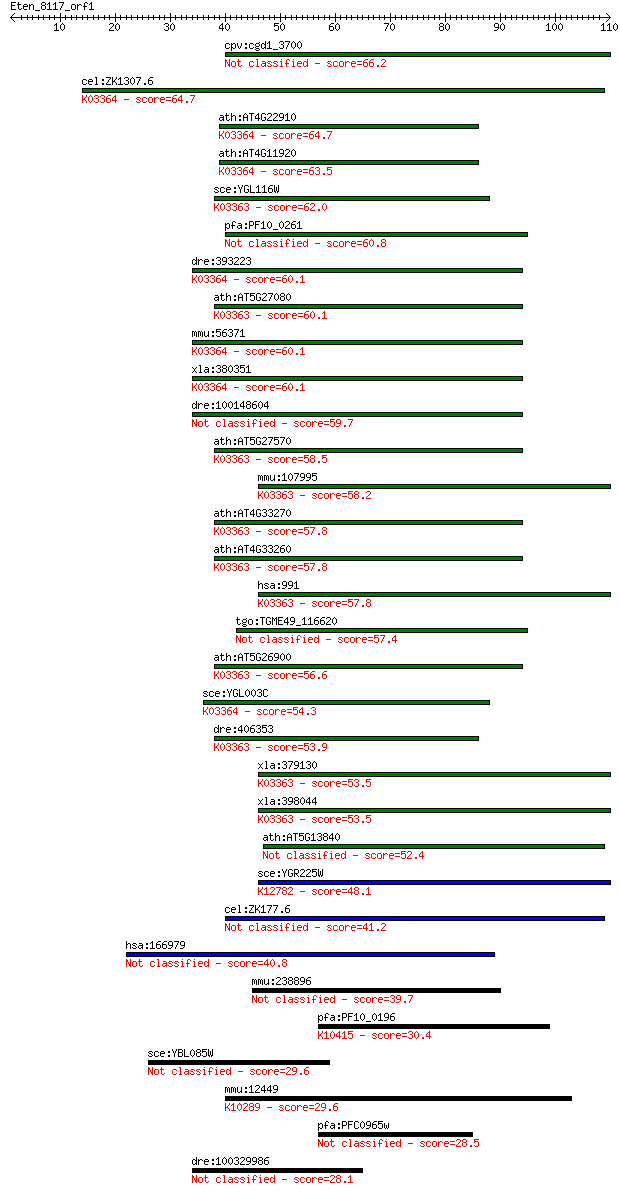
cpv:cgd1_3700 hypothetical protein 66.2 2e-11
cel:ZK1307.6 fzr-1; FiZzy Related family member (fzr-1); K0336... 64.7 7e-11
ath:AT4G22910 FZR2; FZR2 (FIZZY-RELATED 2); signal transducer;... 64.7 7e-11
ath:AT4G11920 CCS52A2; signal transducer; K03364 cell division... 63.5 1e-10
sce:YGL116W CDC20, PAC5; Cdc20p; K03363 cell division cycle 20... 62.0 4e-10
pfa:PF10_0261 WD-repeat protein, putative 60.8 1e-09
dre:393223 fzr1, MGC55450, zgc:55450; fizzy/cell division cycl... 60.1 2e-09
ath:AT5G27080 WD-40 repeat family protein; K03363 cell divisio... 60.1 2e-09
mmu:56371 Fzr1, AW108046, Cdh1, FZR, FZR2, Fyr, HCDH, HCDH1; f... 60.1 2e-09
xla:380351 fzr1, MGC53225; fizzy/cell division cycle 20 relate... 60.1 2e-09
dre:100148604 Fizzy-related protein homolog 59.7 2e-09
ath:AT5G27570 WD-40 repeat family protein; K03363 cell divisio... 58.5 4e-09
mmu:107995 Cdc20, 2310042N09Rik, C87100, p55CDC; cell division... 58.2 7e-09
ath:AT4G33270 CDC20.1; signal transducer; K03363 cell division... 57.8 8e-09
ath:AT4G33260 CDC20.2; signal transducer; K03363 cell division... 57.8 9e-09
hsa:991 CDC20, CDC20A, MGC102824, bA276H19.3, p55CDC; cell div... 57.8 9e-09
tgo:TGME49_116620 WD-40 repeats-containing protein 57.4 1e-08
ath:AT5G26900 WD-40 repeat family protein; K03363 cell divisio... 56.6 2e-08
sce:YGL003C CDH1, HCT1; Cdh1p; K03364 cell division cycle 20-l... 54.3 1e-07
dre:406353 cdc20, wu:fe49f10, zgc:63536; cell division cycle 2... 53.9 1e-07
xla:379130 cdc20; cell division cycle 20 homolog; K03363 cell ... 53.5 2e-07
xla:398044 cdc20, X-FZY; cell division cycle 20 homolog; K0336... 53.5 2e-07
ath:AT5G13840 FZR3; FZR3 (FIZZY-RELATED 3); signal transducer 52.4 3e-07
sce:YGR225W AMA1, SPO70; Activator of meiotic anaphase promoti... 48.1 7e-06
cel:ZK177.6 fzy-1; FiZzY (CDC20 protein family) homolog family... 41.2 9e-04
hsa:166979 CDC20B, FLJ37927; cell division cycle 20 homolog B ... 40.8 0.001
mmu:238896 Cdc20b, EG238896, EG622422; cell division cycle 20 ... 39.7 0.002
pfa:PF10_0196 cytoplasmic dynein intermediate chain, putative;... 30.4 1.5
sce:YBL085W BOI1, BOB1, GIN7; Boi1p 29.6 2.4
mmu:12449 Ccnf, CycF, Fbxo1; cyclin F; K10289 F-box protein 1 ... 29.6 2.8
pfa:PFC0965w conserved Plasmodium protein, unknown function 28.5 6.1
dre:100329986 hypothetical protein LOC100329986 28.1 6.7
> cpv:cgd1_3700 hypothetical protein
Length=453
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/70 (45%), Positives = 47/70 (67%), Gaps = 3/70 (4%)
Query 40 QRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNP 99
+++ K P+++L+ P L DDFYLNL+ WSS N LAVG+ +S+ LW+A T L LN
Sbjct 128 RKIPKGPFKILDAPNLQDDFYLNLVDWSSTNLLAVGLSSSVYLWSASTCKVTNL---LNL 184
Query 100 QPNEEVSAVS 109
Q + V++VS
Sbjct 185 QDQDTVTSVS 194
> cel:ZK1307.6 fzr-1; FiZzy Related family member (fzr-1); K03364
cell division cycle 20-like protein 1, cofactor of APC complex
Length=702
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 56/96 (58%), Gaps = 3/96 (3%)
Query 14 CSNSPFCCCLDCIEMRVIEKRSGGKP-QRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFL 72
+ SPF R++ R+ KP ++V KNPY+VL+ P+L DDFYLNL+ WSS N L
Sbjct 351 TATSPFGGPFGVDSQRLL--RTPRKPIRKVPKNPYKVLDAPELQDDFYLNLVDWSSQNQL 408
Query 73 AVGIQNSLLLWNADTNATRPLFHRLNPQPNEEVSAV 108
+VG+ + LW+A T+ L ++V++V
Sbjct 409 SVGLAACVYLWSATTSQVIKLCDLGQTNEQDQVTSV 444
> ath:AT4G22910 FZR2; FZR2 (FIZZY-RELATED 2); signal transducer;
K03364 cell division cycle 20-like protein 1, cofactor of
APC complex
Length=483
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 27/47 (57%), Positives = 37/47 (78%), Gaps = 0/47 (0%)
Query 39 PQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNA 85
P++V ++PY+VL+ P L DDFYLNL+ WS+ N LAVG+ N + LWNA
Sbjct 157 PRKVPRSPYKVLDAPALQDDFYLNLVDWSAQNVLAVGLGNCVYLWNA 203
> ath:AT4G11920 CCS52A2; signal transducer; K03364 cell division
cycle 20-like protein 1, cofactor of APC complex
Length=475
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/47 (55%), Positives = 37/47 (78%), Gaps = 0/47 (0%)
Query 39 PQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNA 85
P+++ ++PY+VL+ P L DDFYLNL+ WS+ N LAVG+ N + LWNA
Sbjct 149 PRKILRSPYKVLDAPALQDDFYLNLVDWSAQNVLAVGLGNCVYLWNA 195
> sce:YGL116W CDC20, PAC5; Cdc20p; K03363 cell division cycle
20, cofactor of APC complex
Length=610
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 26/50 (52%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 38 KPQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADT 87
K ++++ NP R+L+ P DDFYLNLLSWS N LA+ + +L LWNA T
Sbjct 241 KLRKINTNPERILDAPGFQDDFYLNLLSWSKKNVLAIALDTALYLWNATT 290
> pfa:PF10_0261 WD-repeat protein, putative
Length=603
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 25/55 (45%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 40 QRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLF 94
+++ PYR+L P+L DDFYLNLL WS N +A + + L LWN +T + LF
Sbjct 224 RKIPNMPYRILSAPELMDDFYLNLLDWSKKNIIATALCDKLYLWNNNTCTNQKLF 278
> dre:393223 fzr1, MGC55450, zgc:55450; fizzy/cell division cycle
20 related 1 (Drosophila); K03364 cell division cycle 20-like
protein 1, cofactor of APC complex
Length=495
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 34 RSGGKPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRP 92
RS KP R +SK P++VL+ P+L DDFYLNL+ WSS N L+VG+ + LW+A T+
Sbjct 164 RSPRKPTRKISKIPFKVLDAPELQDDFYLNLVDWSSLNVLSVGLGTCVYLWSACTSQVTR 223
Query 93 L 93
L
Sbjct 224 L 224
> ath:AT5G27080 WD-40 repeat family protein; K03363 cell division
cycle 20, cofactor of APC complex
Length=466
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 40/57 (70%), Gaps = 1/57 (1%)
Query 38 KPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPL 93
KP+R + +N RVL+ P L DDFYLNLL W SAN LA+ + +++ LW+A + +T L
Sbjct 102 KPRRYIPQNSERVLDAPGLMDDFYLNLLDWGSANVLAIALGDTVYLWDASSGSTSEL 158
> mmu:56371 Fzr1, AW108046, Cdh1, FZR, FZR2, Fyr, HCDH, HCDH1;
fizzy/cell division cycle 20 related 1 (Drosophila); K03364
cell division cycle 20-like protein 1, cofactor of APC complex
Length=493
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 34 RSGGKPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRP 92
RS KP R +SK P++VL+ P+L DDFYLNL+ WSS N L+VG+ + LW+A T+
Sbjct 162 RSPRKPTRKISKIPFKVLDAPELQDDFYLNLVDWSSLNVLSVGLGTCVYLWSACTSQVTR 221
Query 93 L 93
L
Sbjct 222 L 222
> xla:380351 fzr1, MGC53225; fizzy/cell division cycle 20 related
1; K03364 cell division cycle 20-like protein 1, cofactor
of APC complex
Length=493
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 34 RSGGKPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRP 92
RS KP R +SK P++VL+ P+L DDFYLNL+ WSS N L+VG+ + LW+A T+
Sbjct 162 RSPRKPTRKISKIPFKVLDAPELQDDFYLNLVDWSSLNVLSVGLGTCVYLWSACTSQVTR 221
Query 93 L 93
L
Sbjct 222 L 222
> dre:100148604 Fizzy-related protein homolog
Length=489
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/61 (49%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 34 RSGGKPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRP 92
RS KP R +SK P++VL+ P+L DDFYLNL+ WS+ N L+VG+ + LW+A T+
Sbjct 159 RSPRKPARKISKIPFKVLDAPELQDDFYLNLVDWSAGNLLSVGLGACVYLWSACTSQVTR 218
Query 93 L 93
L
Sbjct 219 L 219
> ath:AT5G27570 WD-40 repeat family protein; K03363 cell division
cycle 20, cofactor of APC complex
Length=411
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 40/57 (70%), Gaps = 1/57 (1%)
Query 38 KPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPL 93
KP+R + +N RVL+ P + DDFYLNLL W S+N LA+ + +++ LW+A + +T L
Sbjct 71 KPRRYIPQNSERVLDAPGIADDFYLNLLDWGSSNVLAIALGDTVYLWDASSGSTYKL 127
> mmu:107995 Cdc20, 2310042N09Rik, C87100, p55CDC; cell division
cycle 20 homolog (S. cerevisiae); K03363 cell division cycle
20, cofactor of APC complex
Length=499
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 46 PYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEEV 105
P R+L+ P++ +D+YLNL+ WSS N LAV + NS+ LWNA + L QP + +
Sbjct 172 PDRILDAPEIRNDYYLNLVDWSSGNVLAVALDNSVYLWNAGSGDILQLLQM--EQPGDYI 229
Query 106 SAVS 109
S+V+
Sbjct 230 SSVA 233
> ath:AT4G33270 CDC20.1; signal transducer; K03363 cell division
cycle 20, cofactor of APC complex
Length=457
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 38 KPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPL 93
KP+R + + R L+ P + DDFYLNLL W SAN LA+ + +++ LW+A T +T L
Sbjct 119 KPRRYIPQTSERTLDAPDIVDDFYLNLLDWGSANVLAIALDHTVYLWDASTGSTSEL 175
> ath:AT4G33260 CDC20.2; signal transducer; K03363 cell division
cycle 20, cofactor of APC complex
Length=447
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 38 KPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPL 93
KP+R + + R L+ P + DDFYLNLL W SAN LA+ + +++ LW+A T +T L
Sbjct 109 KPRRYIPQTSERTLDAPDIVDDFYLNLLDWGSANVLAIALDHTVYLWDASTGSTSEL 165
> hsa:991 CDC20, CDC20A, MGC102824, bA276H19.3, p55CDC; cell division
cycle 20 homolog (S. cerevisiae); K03363 cell division
cycle 20, cofactor of APC complex
Length=499
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 46 PYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEEV 105
P R+L+ P++ +D+YLNL+ WSS N LAV + NS+ LW+A + L QP E +
Sbjct 172 PDRILDAPEIRNDYYLNLVDWSSGNVLAVALDNSVYLWSASSGDILQLLQM--EQPGEYI 229
Query 106 SAVS 109
S+V+
Sbjct 230 SSVA 233
> tgo:TGME49_116620 WD-40 repeats-containing protein
Length=1218
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 35/59 (59%), Gaps = 6/59 (10%)
Query 42 VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWN------ADTNATRPLF 94
V + PYR L P L DDFYLNL+ WS AN LAV +++ L LW+ AD R LF
Sbjct 357 VPQRPYRTLPAPDLLDDFYLNLVDWSRANLLAVALKSKLFLWSPQPRHFADGRQARLLF 415
> ath:AT5G26900 WD-40 repeat family protein; K03363 cell division
cycle 20, cofactor of APC complex
Length=444
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 38 KPQR-VSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPL 93
KP+R + +N RVL+ P L DDF LNLL W SAN LA+ + +++ LW+A + +T L
Sbjct 105 KPRRYIPQNSERVLDAPGLRDDFSLNLLDWGSANVLAIALGDTVYLWDASSGSTSEL 161
> sce:YGL003C CDH1, HCT1; Cdh1p; K03364 cell division cycle 20-like
protein 1, cofactor of APC complex
Length=566
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 36/52 (69%), Gaps = 0/52 (0%)
Query 36 GGKPQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADT 87
G + ++++K PYRVL+ P L DDFY +L+ WSS + LAV + S+ L + +T
Sbjct 241 GKQFRQIAKVPYRVLDAPSLADDFYYSLIDWSSTDVLAVALGKSIFLTDNNT 292
> dre:406353 cdc20, wu:fe49f10, zgc:63536; cell division cycle
20 homolog; K03363 cell division cycle 20, cofactor of APC
complex
Length=496
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 38 KPQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNA 85
K + +S P R+L+ P + +DFYLNL+ W N LAVG+ N + LW+A
Sbjct 165 KSRYISSYPERILDAPDIRNDFYLNLMDWGRQNVLAVGLANQVYLWDA 212
> xla:379130 cdc20; cell division cycle 20 homolog; K03363 cell
division cycle 20, cofactor of APC complex
Length=506
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 2/64 (3%)
Query 46 PYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEEV 105
P RVL+ P + +D+YLNL+ WSS N LAV + +S+ LWN T L N + E +
Sbjct 179 PDRVLDAPDIRNDYYLNLIDWSSQNALAVALNDSVYLWNYATGDIILLLQMENSE--EYI 236
Query 106 SAVS 109
S+VS
Sbjct 237 SSVS 240
> xla:398044 cdc20, X-FZY; cell division cycle 20 homolog; K03363
cell division cycle 20, cofactor of APC complex
Length=507
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 2/64 (3%)
Query 46 PYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEEV 105
P RVL+ P + +D+YLNL+ WSS N LAV + +S+ LWN T L N + E +
Sbjct 180 PDRVLDAPDIRNDYYLNLIDWSSQNALAVALNDSVYLWNYATGDIILLLQMENSE--EYI 237
Query 106 SAVS 109
S+VS
Sbjct 238 SSVS 241
> ath:AT5G13840 FZR3; FZR3 (FIZZY-RELATED 3); signal transducer
Length=481
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 37/62 (59%), Gaps = 3/62 (4%)
Query 47 YRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEEVS 106
++VL+ P L DDFYLN++ WSS N LAVG+ + LW A + L + PN+ V
Sbjct 163 HKVLDAPSLQDDFYLNVVDWSSQNVLAVGLGTCVYLWTASNSKVTKL---CDLGPNDSVC 219
Query 107 AV 108
+V
Sbjct 220 SV 221
> sce:YGR225W AMA1, SPO70; Activator of meiotic anaphase promoting
complex (APC/C); Cdc20p family member; required for initiation
of spore wall assembly; required for Clb1p degradation
during meiosis; K12782 meiosis-specific APC/C activator protein
AMA1
Length=593
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Query 46 PYRVLEVPQLPDDFYLNLLSWS-SANFLAVGIQNSLLLWNADTNATRPLFHRLNPQPNEE 104
PYRVL+ P L +DFY NL+SWS + N + VG+ S+ +W+ A L H+ + +
Sbjct 216 PYRVLDAPCLRNDFYSNLISWSRTTNNVLVGLGCSVYIWSEKEGAVSILDHQYLSEKRDL 275
Query 105 VSAVS 109
V+ VS
Sbjct 276 VTCVS 280
> cel:ZK177.6 fzy-1; FiZzY (CDC20 protein family) homolog family
member (fzy-1)
Length=507
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 34/69 (49%), Gaps = 0/69 (0%)
Query 40 QRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNATRPLFHRLNP 99
+ V + +VL+ P L D Y L W N++AV + + L LWN +T + LF P
Sbjct 159 RHVKETATKVLDGPGLTKDLYSRHLDWGCHNWVAVALGHELYLWNTETCVIKNLFEDNAP 218
Query 100 QPNEEVSAV 108
+++V
Sbjct 219 TNEGLITSV 227
> hsa:166979 CDC20B, FLJ37927; cell division cycle 20 homolog
B (S. cerevisiae)
Length=477
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 22 CLDCIEMRVIEKRSGGKPQRVSKNPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLL 81
C D + +S G P + + L +D+YLN+L WS N +A+ + +++
Sbjct 195 CKDGVRDESFHLKSSGDINDSILQPEVKIHITGLRNDYYLNILDWSFQNLVAIALGSAVY 254
Query 82 LWNADTN 88
+WN + +
Sbjct 255 IWNGENH 261
> mmu:238896 Cdc20b, EG238896, EG622422; cell division cycle 20
homolog B (S. cerevisiae)
Length=536
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 45 NPYRVLEVPQLPDDFYLNLLSWSSANFLAVGIQNSLLLWNADTNA 89
P + + L +D+YLN L WSS N +AV + S+ +WN ++
Sbjct 266 QPEVKIHLTGLRNDYYLNTLDWSSQNLVAVALGTSVYIWNGQNHS 310
> pfa:PF10_0196 cytoplasmic dynein intermediate chain, putative;
K10415 dynein intermediate chain, cytosolic
Length=769
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 2/44 (4%)
Query 57 DDFYLNLLSWS--SANFLAVGIQNSLLLWNADTNATRPLFHRLN 98
D+ N L+WS +A Q L LWNA + +P F LN
Sbjct 703 DNSSTNKLAWSHDGKRLIAADTQGCLTLWNASSEIYQPKFEDLN 746
> sce:YBL085W BOI1, BOB1, GIN7; Boi1p
Length=980
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 22/35 (62%), Gaps = 2/35 (5%)
Query 26 IEMRVIEKRSGGKPQRVSKNPYRVLEV--PQLPDD 58
I+ + E RSG Q VSK+P RV EV PQL D+
Sbjct 130 IDKALEELRSGSVEQEVSKSPTRVPEVSTPQLQDE 164
> mmu:12449 Ccnf, CycF, Fbxo1; cyclin F; K10289 F-box protein
1 (cyclin F)
Length=777
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 5/67 (7%)
Query 40 QRVSKNPYRVLEVPQLPDDFYLNLLSW-SSANFLAVGIQNSLLLWNADTNAT---RPLFH 95
+R+ + P R L + LP+D ++L W S + LAV +S L + D +A+ F
Sbjct 21 RRIKRRP-RNLTILSLPEDVLFHILKWLSVGDILAVRAVHSHLKYLVDNHASVWASASFQ 79
Query 96 RLNPQPN 102
L P P
Sbjct 80 ELWPSPQ 86
> pfa:PFC0965w conserved Plasmodium protein, unknown function
Length=2706
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 57 DDFYLNLLSWSSANFLAVGIQNSLLLWN 84
++ +L L S +SANF+ V S+ +WN
Sbjct 847 EEIFLFLFSNTSANFITVQTNGSIYMWN 874
> dre:100329986 hypothetical protein LOC100329986
Length=168
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 34 RSGGKPQRVSKNPYRVLEVPQLPDDFYLNLL 64
++ G PQRVS + V ++P++ DDF N L
Sbjct 60 QTDGSPQRVSSARHPVPKIPEVVDDFLRNYL 90
Lambda K H
0.320 0.136 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40