bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8049_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
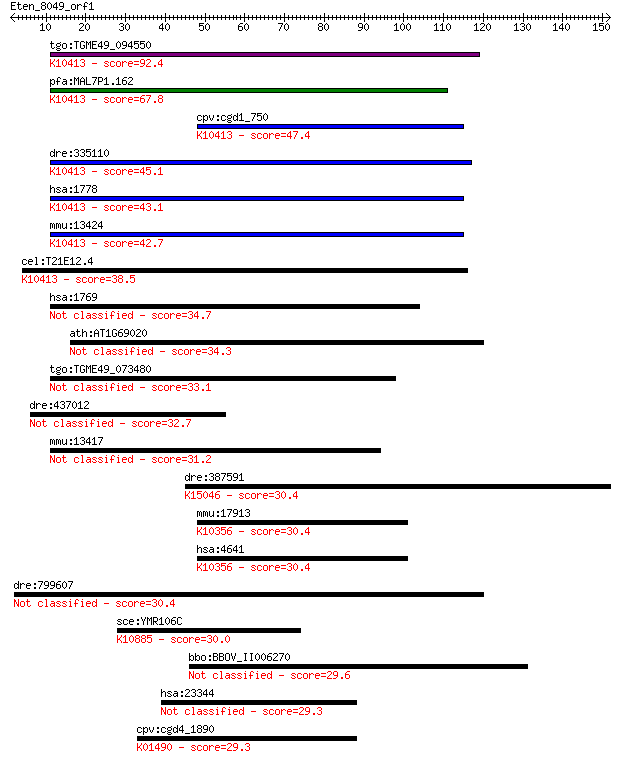
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 92.4 5e-19
pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein hea... 67.8 1e-11
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 47.4 2e-05
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 45.1 9e-05
hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNE... 43.1 4e-04
mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,... 42.7 4e-04
cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1); ... 38.5 0.009
hsa:1769 DNAH8, ATPase, FLJ25850, FLJ36115, FLJ36334, hdhc9; d... 34.7 0.11
ath:AT1G69020 prolyl oligopeptidase family protein 34.3 0.14
tgo:TGME49_073480 axonemal beta dynein heavy chain, putative (... 33.1 0.34
dre:437012 zgc:100980 32.7 0.47
mmu:13417 Dnahc8, ATPase, Hst6.7b, P1-Loop; dynein, axonemal, ... 31.2 1.3
dre:387591 ivns1abpa, cb1052, fj23g11, ivns1abp, wu:fj23g11; i... 30.4 2.1
mmu:17913 Myo1c, C80397, MMIb, MYO1E, NMI, mm1beta, myr2; myos... 30.4 2.1
hsa:4641 MYO1C, FLJ23903, MMI-beta, MMIb, NMI, myr2; myosin IC... 30.4 2.4
dre:799607 proto-oncogene serine/threonine-protein kinase Pim-... 30.4 2.4
sce:YMR106C YKU80, HDF2; Subunit of the telomeric Ku complex (... 30.0 2.8
bbo:BBOV_II006270 18.m10022; hypothetical protein 29.6 3.5
hsa:23344 ESYT1, FAM62A, KIAA0747, MBC2; extended synaptotagmi... 29.3 4.9
cpv:cgd4_1890 adenosine monophosphate deaminase 2 ; K01490 AMP... 29.3 5.1
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 71/112 (63%), Gaps = 6/112 (5%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNM----PNLGKDVEDSEAELLRAPAELFKTHEQYLEWV 66
++L S VDYLF+P AFE+ F LN+ G D S +LL AP +LFK Y W
Sbjct 4485 KILSSLVDYLFQPAAFEASFPLNIMAENEETGAD-SLSLPQLLTAP-DLFKQASAYEAWA 4542
Query 67 DSLPPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEESLDFHSIS 118
+ L D P W+GF+++AE LLASRQ+ A + +W+ +LLRG EE+ DF +IS
Sbjct 4543 EQLWSVDSPTWLGFSAQAERLLASRQSLAAVGSWAAVLLRGNEEAHDFQAIS 4594
> pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein heavy
chain 1, cytosolic
Length=4985
Score = 67.8 bits (164), Expect = 1e-11, Method: Composition-based stats.
Identities = 34/103 (33%), Positives = 59/103 (57%), Gaps = 11/103 (10%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPN---LGKDVEDSEAELLRAPAELFKTHEQYLEWVD 67
++L +F+D+L +FE+DF LN+ N L KD L +P +LF+ Y+ W +
Sbjct 4554 KILDTFIDHLMNSNSFETDFKLNICNSTSLNKD-------FLVSP-DLFRNINDYINWTN 4605
Query 68 SLPPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEE 110
++ D+PAW+GF +AE LL +R + I+ W+ L + + +
Sbjct 4606 NMSNTDLPAWLGFGQQAEGLLTTRTNFSIISKWNILYSKSRSD 4648
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 48 LLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRG 107
+L AP + K + YL+WV+ L + P W+G + AE ++++++ NWS L++R
Sbjct 4801 ILLAP-DTSKKVQNYLDWVEKLSSTNWPTWLGLSPLAENVISAQKGQKMSLNWSVLIIRS 4859
Query 108 KEESLDF 114
+ E+ D
Sbjct 4860 RNETPDL 4866
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 55/107 (51%), Gaps = 11/107 (10%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLG-KDVEDSEAELLRAPAELFKTHEQYLEWVDSL 69
R+L +F++ LF +F+S+F L + G KD+ + P + + E+++ WV+ L
Sbjct 4262 RLLNTFLERLFTCSSFDSEFKLALKVDGHKDI--------KMPDGIRR--EEFIHWVEML 4311
Query 70 PPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEESLDFHS 116
P P+W+G S AE +L + Q +A + + E+ L + +
Sbjct 4312 PDTQTPSWLGLPSNAEKVLLTTQGTDMMAKLLKMQMLEDEDDLAYET 4358
> hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNECL,
DYHC, Dnchc1, HL-3, KIAA0325, p22; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4646
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 54/105 (51%), Gaps = 11/105 (10%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLG-KDVEDSEAELLRAPAELFKTHEQYLEWVDSL 69
R+L +F++ LF +F+S+F L G KD++ + +R E++++WV+ L
Sbjct 4263 RLLNTFLERLFTTRSFDSEFKLACKVDGHKDIQMPDG--IR--------REEFVQWVELL 4312
Query 70 PPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEESLDF 114
P P+W+G + AE +L + Q I+ + + E+ L +
Sbjct 4313 PDTQTPSWLGLPNNAERVLLTTQGVDMISKMLKMQMLEDEDDLAY 4357
> mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,
Dnchc1, Dnec1, Dnecl, Loa, MAP1C, P22, Swl, mKIAA0325; dynein
cytoplasmic 1 heavy chain 1; K10413 dynein heavy chain
1, cytosolic
Length=4644
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 54/105 (51%), Gaps = 11/105 (10%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLG-KDVEDSEAELLRAPAELFKTHEQYLEWVDSL 69
R+L +F++ LF +F+S+F L G KD++ + +R E++++WV+ L
Sbjct 4261 RLLNTFLERLFTTRSFDSEFKLACKVDGHKDIQMPDG--IR--------REEFVQWVELL 4310
Query 70 PPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEESLDF 114
P P+W+G + AE +L + Q I+ + + E+ L +
Sbjct 4311 PDAQTPSWLGLPNNAERVLLTTQGVDMISKMLKMQMLEDEDDLAY 4355
> cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1);
K10413 dynein heavy chain 1, cytosolic
Length=4568
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 53/113 (46%), Gaps = 25/113 (22%)
Query 4 NCVVTCIRVLRSF-VDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQY 62
+CV+ + +SF D++ P+ ++ D L PN+ K +Q
Sbjct 4214 DCVLENLFTAKSFEQDHVLIPK-YDGDDSLFTPNMSK-------------------KDQM 4253
Query 63 LEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEESLDFH 115
+ WV+ L +PAW+G + AE +L +++ + + N +L+ +E L F+
Sbjct 4254 IGWVEELKNEQLPAWLGLPNNAEKVLLTKRGESMLRN----MLKVTDEELAFN 4302
> hsa:1769 DNAH8, ATPase, FLJ25850, FLJ36115, FLJ36334, hdhc9;
dynein, axonemal, heavy chain 8
Length=4490
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 36/93 (38%), Gaps = 15/93 (16%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQYLEWVDSLP 70
R+L F F + FE FC + P L KT +QY E++ SLP
Sbjct 4127 RLLNCFARVWFSEKMFEPSFCFYTG-------------YKIP--LCKTLDQYFEYIQSLP 4171
Query 71 PRDMPAWIGFNSRAEWLLASRQAHACIANWSTL 103
D P G + A+ S A A + + +
Sbjct 4172 SLDNPEVFGLHPNADITYQSNTASAVLETITNI 4204
> ath:AT1G69020 prolyl oligopeptidase family protein
Length=757
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 52/106 (49%), Gaps = 13/106 (12%)
Query 16 FVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQYLEWVDSLPPRDMP 75
FVD+L R ++ F + L +D+ SE + R P E+F E++ +W L + +P
Sbjct 72 FVDFLKRENSYSQAFMADTETLRRDLF-SEMKT-RIPEEIFTPPERWGQW---LYRQYIP 126
Query 76 AWIGFNSRAEWLLASRQAHACIANWSTLLLRGKEES--LDFHSISE 119
E+ L R+ NW + L RG+EE LD++ I+E
Sbjct 127 ------KGKEYPLLCRRLEKGKTNWLSGLFRGEEEEVVLDWNQIAE 166
> tgo:TGME49_073480 axonemal beta dynein heavy chain, putative
(EC:5.99.1.3)
Length=4273
Score = 33.1 bits (74), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 39/87 (44%), Gaps = 12/87 (13%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQYLEWVDSLP 70
R++ +D ++ PE E FCL S +E + P F T E+YL+++ +P
Sbjct 3860 RLINYLIDDIYSPEILEEGFCL-----------SASEGIEVPPATF-TLEEYLDFIREMP 3907
Query 71 PRDMPAWIGFNSRAEWLLASRQAHACI 97
+ P + A +A +A + +
Sbjct 3908 TEESPEVYTLHPNANMSVAISEASSIL 3934
> dre:437012 zgc:100980
Length=804
Score = 32.7 bits (73), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 6 VVTCIRVLRSFVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAE 54
V+ C R+L + Y+F + F +P LGK V++ E E R AE
Sbjct 94 VLICTRLLTRILPYIFEDPDWHGFFWSTVPKLGKSVDEGEEECTRPLAE 142
> mmu:13417 Dnahc8, ATPase, Hst6.7b, P1-Loop; dynein, axonemal,
heavy chain 8
Length=4731
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 32/83 (38%), Gaps = 15/83 (18%)
Query 11 RVLRSFVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQYLEWVDSLP 70
R+L F F + FE FC + P + KT +QY E++ SLP
Sbjct 4368 RLLNCFARVWFSEKMFEPSFCFYTG-------------YKIP--ICKTLDQYFEFIQSLP 4412
Query 71 PRDMPAWIGFNSRAEWLLASRQA 93
D P G + A+ S A
Sbjct 4413 SLDNPEVFGLHPNADITYQSNTA 4435
> dre:387591 ivns1abpa, cb1052, fj23g11, ivns1abp, wu:fj23g11;
influenza virus NS1A binding protein a; K15046 influenza virus
NS1A-binding protein
Length=643
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 44/107 (41%), Gaps = 6/107 (5%)
Query 45 EAELLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANWSTLL 104
+ LL AE+F T + ++++V PPR+ S + L H C W +
Sbjct 233 DGSLLDGQAEVFGTEDDHIQFVQKKPPRENVHRQLSTSSSGSLSPGSTKHPCKQEWKYI- 291
Query 105 LRGKEESLDFHSISESIMKGLKRQKSVAAISDEKEPDEAPKATSWLT 151
E++ + + +++ G+ V + P +P AT LT
Sbjct 292 --ASEKTTNNTYLCLAVLNGM---LCVIFLHGRSSPQASPSATPCLT 333
> mmu:17913 Myo1c, C80397, MMIb, MYO1E, NMI, mm1beta, myr2; myosin
IC; K10356 myosin I
Length=1028
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 48 LLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANW 100
+R P LF T + SL + AW GF+ R ++L R A CI +W
Sbjct 681 FIRFPKTLFATEDSLEVRRQSLATKIQAAWRGFHWRQKFLRVKRSA-ICIQSW 732
> hsa:4641 MYO1C, FLJ23903, MMI-beta, MMIb, NMI, myr2; myosin
IC; K10356 myosin I
Length=1063
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 48 LLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANW 100
+R P LF T + SL + AW GF+ R ++L R A CI +W
Sbjct 716 FIRFPKTLFATEDALEVRRQSLATKIQAAWRGFHWRQKFLRVKRSA-ICIQSW 767
> dre:799607 proto-oncogene serine/threonine-protein kinase Pim-1-like
Length=244
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 29/123 (23%), Positives = 47/123 (38%), Gaps = 9/123 (7%)
Query 2 TANCVVTCIRVLRSFVDYLFRPEAFESDFCLNMPNLGKDVEDSEAELLRAPAELFKTHEQ 61
TAN +VT R+ +D+ + F N + E LR +F +
Sbjct 107 TANILVTLPRLKLKLIDFGCAQPITQKPF--NKSDYRGAGHGMPPEALRC--RVFHANPA 162
Query 62 YLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAH-----ACIANWSTLLLRGKEESLDFHS 116
Y+ + + M + FN+R + S Q H AC+ S L+R + L H
Sbjct 163 YVWTIGIMLYEIMHGRLAFNNRQSIMFGSVQIHPRLSTACVDLTSQCLIRNPAKRLQLHQ 222
Query 117 ISE 119
+ E
Sbjct 223 VEE 225
> sce:YMR106C YKU80, HDF2; Subunit of the telomeric Ku complex
(Yku70p-Yku80p), involved in telomere length maintenance, structure
and telomere position effect; relocates to sites of
double-strand cleavage to promote nonhomologous end joining
during DSB repair; K10885 ATP-dependent DNA helicase 2 subunit
2
Length=629
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 28 SDFCLNMPNLGKDVEDSEAELLRAPAE-LFKTHEQYLEWVDSLPPRD 73
+D L +P+L KD E+++ + LR P +++ + LEW+ L D
Sbjct 503 TDTTLPLPSLNKDQEENKKDPLRIPTVFVYRQQQVLLEWIHQLMIND 549
> bbo:BBOV_II006270 18.m10022; hypothetical protein
Length=1171
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 28/89 (31%), Positives = 38/89 (42%), Gaps = 12/89 (13%)
Query 46 AELLRAPAELF----KTHEQYLEWVDSLPPRDMPAWIGFNSRAEWLLASRQAHACIANWS 101
+E LR P EL K E +E V PRD W+ R E +L S H +A
Sbjct 757 SEALRTPMELLDGDMKVPETLIEQVLERGPRDNKTWLLLLKRFELILHSF-GHKHLARVL 815
Query 102 TLLLRGKEESLDFHSISESIMKGLKRQKS 130
+L +G + I ++ LKRQ S
Sbjct 816 VMLPKGLQ-------IPPELVPKLKRQAS 837
> hsa:23344 ESYT1, FAM62A, KIAA0747, MBC2; extended synaptotagmin-like
protein 1
Length=1114
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 39 KDVEDSEAELLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNS--RAEWL 87
+ V D + LRA +L EQ + R++PAW+ F +AEWL
Sbjct 92 RRVRDEKERSLRAARQLLDDEEQLTAKTLYMSHRELPAWVSFPDVEKAEWL 142
> cpv:cgd4_1890 adenosine monophosphate deaminase 2 ; K01490 AMP
deaminase [EC:3.5.4.6]
Length=846
Score = 29.3 bits (64), Expect = 5.1, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 6/55 (10%)
Query 33 NMPNLGKDVEDSEAELLRAPAELFKTHEQYLEWVDSLPPRDMPAWIGFNSRAEWL 87
N P GK + + E+++ +L TH Q++EW S+ +D W + AEWL
Sbjct 442 NNPIKGKYLAEITKEVIQ---DLKTTHYQFVEWRISVYGKDKSEW---KTLAEWL 490
Lambda K H
0.318 0.131 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40