bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8042_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
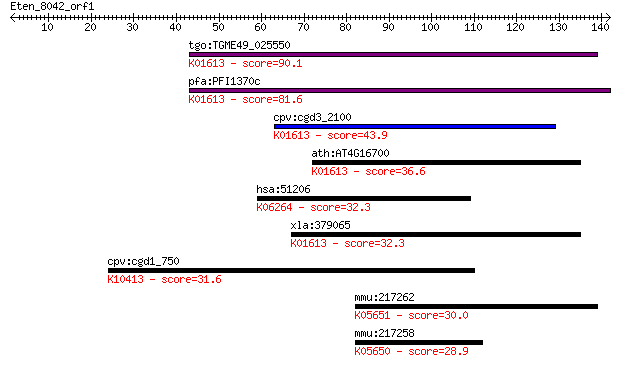
tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme, ... 90.1 2e-18
pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1... 81.6 7e-16
cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosph... 43.9 2e-04
ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);... 36.6 0.027
hsa:51206 GP6, GPIV, GPVI, MGC138168; glycoprotein VI (platele... 32.3 0.42
xla:379065 MGC52759; similar to phosphatidylserine decarboxyla... 32.3 0.51
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 31.6 0.86
mmu:217262 Abca9, D630040K07Rik; ATP-binding cassette, sub-fam... 30.0 2.7
mmu:217258 Abca8a; ATP-binding cassette, sub-family A (ABC1), ... 28.9 5.5
> tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme,
putative (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=337
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 61/96 (63%), Gaps = 0/96 (0%)
Query 43 KFREALFVTGNAADVQSPNKLFCLRLVFGRARSRLLGRLMSVKIPSSLRPLLTTLFLRLQ 102
KFRE + T N A++QSP+KLF LRL+FGR RSR+ G +M++ I +LR + +
Sbjct 16 KFREVIAATDNVAEIQSPSKLFYLRLLFGRTRSRITGSVMNINIMPALRDPIYRTLASVG 75
Query 103 PSAAADSRYPLEAYESFGQLFCRTLRDRAREVMDLS 138
+ RYPL +Y+ G LF RTL+D+ RE+ D+
Sbjct 76 GIDTEEIRYPLRSYKCIGHLFARTLKDKEREIEDIG 111
> pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1.65);
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=353
Score = 81.6 bits (200), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 59/99 (59%), Gaps = 0/99 (0%)
Query 43 KFREALFVTGNAADVQSPNKLFCLRLVFGRARSRLLGRLMSVKIPSSLRPLLTTLFLRLQ 102
K+ E L V + +VQ ++LF RL+FGR RSR+ GR+ +++IP S R + F++
Sbjct 34 KYHEVLTVYEDKTNVQQSSRLFWTRLLFGRTRSRITGRIFNIEIPHSYRLYVYNFFIKYL 93
Query 103 PSAAADSRYPLEAYESFGQLFCRTLRDRAREVMDLSPLA 141
+ +YP+E+Y+S G F R +R+ R + DL+ +
Sbjct 94 NINKEEIKYPIESYKSLGDFFSRYIREDTRPIGDLNEYS 132
> cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=314
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 16/82 (19%)
Query 63 LFCLRLVFGRARSRLLGRLMSVKIPSSLR----PLLTTLFLRLQPSAAADSR-------- 110
+F R +FGR RSR LG+L+++ +P S+R L +L S +A S+
Sbjct 1 IFFYRSLFGRTRSRFLGKLLNINLPVSIRRRIYGFLINNYLYKDSSFSAYSKDEKIKKFE 60
Query 111 ----YPLEAYESFGQLFCRTLR 128
L++Y S G+LF R++R
Sbjct 61 EKHANSLDSYRSIGELFTRSIR 82
> ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=453
Score = 36.6 bits (83), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 0/63 (0%)
Query 72 RARSRLLGRLMSVKIPSSLRPLLTTLFLRLQPSAAADSRYPLEAYESFGQLFCRTLRDRA 131
R+ SR G MS++IP +RP + R S ++ PLE Y S F R+L++
Sbjct 125 RSISRAWGSFMSLEIPVWMRPYAYKAWARAFHSNLEEAALPLEEYTSLQDFFVRSLKEGC 184
Query 132 REV 134
R +
Sbjct 185 RPI 187
> hsa:51206 GP6, GPIV, GPVI, MGC138168; glycoprotein VI (platelet);
K06264 platelet glycoprotein VI
Length=620
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 26/52 (50%), Gaps = 2/52 (3%)
Query 59 SPNKLFCLRLVFGR--ARSRLLGRLMSVKIPSSLRPLLTTLFLRLQPSAAAD 108
SP LFCL L GR A+S L + +PSSL PL + LR Q D
Sbjct 4 SPTALFCLGLCLGRVPAQSGPLPKPSLQALPSSLVPLEKPVTLRCQGPPGVD 55
> xla:379065 MGC52759; similar to phosphatidylserine decarboxylase;
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=355
Score = 32.3 bits (72), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 5/73 (6%)
Query 67 RLVFGRARSRLLGRLMS----VKIPSSLR-PLLTTLFLRLQPSAAADSRYPLEAYESFGQ 121
+ ++ RA +RLL RL V +P LR PLL+ + A L Y++ G+
Sbjct 60 KAMYSRAPTRLLSRLWGLINEVNLPCWLRRPLLSFYVWAFSVNMAEAEEEDLNRYQNLGE 119
Query 122 LFCRTLRDRAREV 134
LF R+L+ AR +
Sbjct 120 LFRRSLKPTARTI 132
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 31.6 bits (70), Expect = 0.86, Method: Composition-based stats.
Identities = 28/88 (31%), Positives = 42/88 (47%), Gaps = 16/88 (18%)
Query 24 RSLGLALATSGCLSWVAVAKFREALFVTGNAADVQSPNKLFCL--RLVFGRARSRLLGRL 81
RS+ L ++G L A+++ +EAL P KL L +L+ S LL +L
Sbjct 2348 RSMKSVLRSAGKLKKSAISENKEAL---------DDPQKLVILEQQLIIRSISSTLLPKL 2398
Query 82 MSVKIPSSLRPLLTTLFLRLQPSAAADS 109
+S +P LLTTLF + P +S
Sbjct 2399 VSTDVP-----LLTTLFQGVFPQVPFES 2421
> mmu:217262 Abca9, D630040K07Rik; ATP-binding cassette, sub-family
A (ABC1), member 9; K05651 ATP-binding cassette, subfamily
A (ABC1), member 9
Length=1623
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 30/65 (46%), Gaps = 11/65 (16%)
Query 82 MSVKIPSSLRPLLTTLFLRLQPSAAADSRY--------PLEAYESFGQLFCRTLRDRARE 133
M VK PS + PL T + +RL P AA RY P+E + F + +R +E
Sbjct 1519 MKVKTPSQVEPLNTEI-MRLFPQAARQERYSSLMVYKLPVEDVRPLSEAFFKL--ERLKE 1575
Query 134 VMDLS 138
DL
Sbjct 1576 NFDLE 1580
> mmu:217258 Abca8a; ATP-binding cassette, sub-family A (ABC1),
member 8a; K05650 ATP-binding cassette, subfamily A (ABC1),
member 8
Length=1619
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 1/30 (3%)
Query 82 MSVKIPSSLRPLLTTLFLRLQPSAAADSRY 111
M VK PS + PL T + +RL P AA RY
Sbjct 1515 MKVKTPSQVEPLNTEI-MRLFPQAARQERY 1543
Lambda K H
0.325 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2618291680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40