bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8015_orf3
Length=191
Score E
Sequences producing significant alignments: (Bits) Value
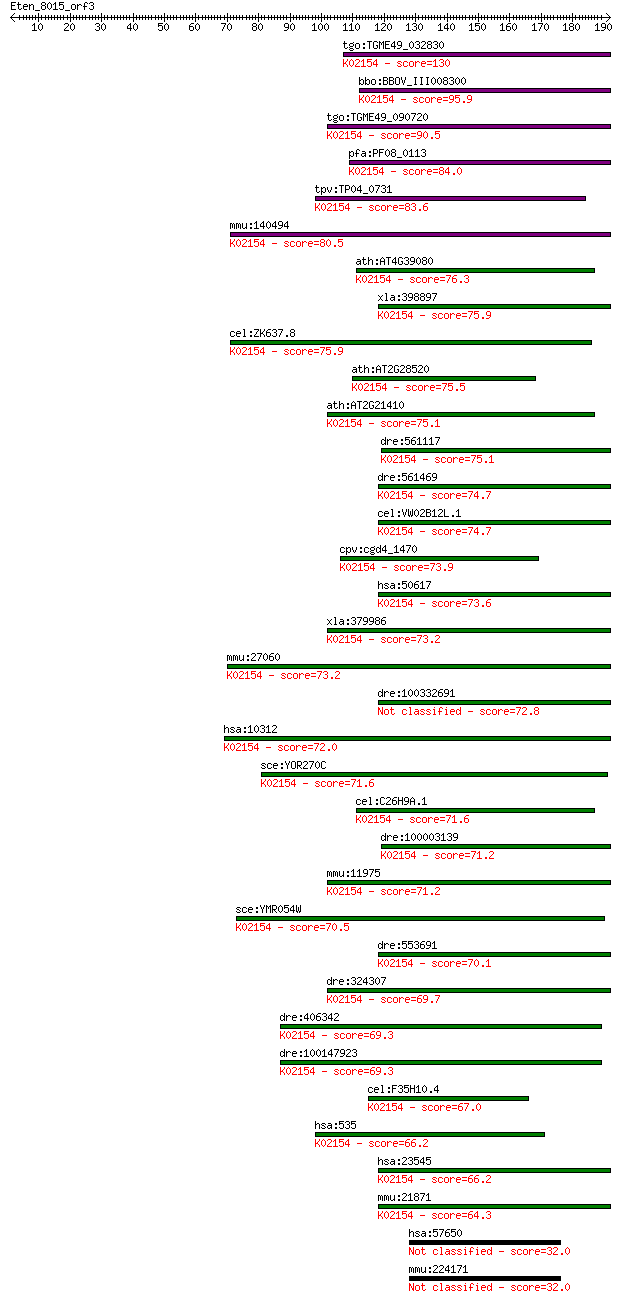
tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit... 130 3e-30
bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit fam... 95.9 7e-20
tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit... 90.5 3e-18
pfa:PF08_0113 vacuolar proton translocating ATPase subunit A, ... 84.0 3e-16
tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14); K... 83.6 4e-16
mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lys... 80.5 4e-15
ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPa... 76.3 6e-14
xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysoso... 75.9 7e-14
cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K021... 75.9 8e-14
ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATP... 75.5 9e-14
ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPa... 75.1 1e-13
dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-tr... 75.1 1e-13
dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-li... 74.7 2e-13
cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);... 74.7 2e-13
cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 tran... 73.9 3e-13
hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,... 73.6 4e-13
xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysoso... 73.2 5e-13
mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,... 73.2 5e-13
dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0 ... 72.8 7e-13
hsa:10312 TCIRG1, ATP6N1C, ATP6V0A3, Atp6i, OC-116kDa, OC116, ... 72.0 1e-12
sce:YOR270C VPH1; Subunit a of vacuolar-ATPase V0 domain, one ... 71.6 1e-12
cel:C26H9A.1 vha-7; Vacuolar H ATPase family member (vha-7); K... 71.6 2e-12
dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator... 71.2 2e-12
mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1... 71.2 2e-12
sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transp... 70.5 4e-12
dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPas... 70.1 5e-12
dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,... 69.7 6e-12
dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154 V... 69.3 7e-12
dre:100147923 T-cell immune regulator 1-like; K02154 V-type H+... 69.3 8e-12
cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5); K... 67.0 4e-11
hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1, ... 66.2 6e-11
hsa:23545 ATP6V0A2, A2, ARCL, ATP6A2, ATP6N1D, J6B7, RTF, STV1... 66.2 6e-11
mmu:21871 Atp6v0a2, 8430408C20Rik, AI385560, ATP6a2, AW489264,... 64.3 3e-10
hsa:57650 CIP2A, FLJ12850, MGC163436, p90; KIAA1524 32.0 1.2
mmu:224171 C330027C09Rik, AA408511, AU018569, Cip2a, Kiaa1524;... 32.0 1.3
> tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=909
Score = 130 bits (326), Expect = 3e-30, Method: Composition-based stats.
Identities = 60/85 (70%), Positives = 76/85 (89%), Gaps = 0/85 (0%)
Query 107 QQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVR 166
+++ EEHEG GD+FI QMIETIEF+LGT+SNTASYLRLWALSLAHQQL+LVFY+QTVVR
Sbjct 773 KEEIGEEHEGPGDIFIHQMIETIEFILGTISNTASYLRLWALSLAHQQLALVFYTQTVVR 832
Query 167 ALTLSEDTAVVAAALFLVFSLFACI 191
A+ L+++T VA ALF++F+ +ACI
Sbjct 833 AIELTDNTTFVALALFVIFAAYACI 857
> bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit family
protein; K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=927
Score = 95.9 bits (237), Expect = 7e-20, Method: Composition-based stats.
Identities = 51/85 (60%), Positives = 63/85 (74%), Gaps = 7/85 (8%)
Query 112 EEHEGA---GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRAL 168
E+H G D+FI Q+IETIEF LG +SNTASYLRLWALSL+HQQLS VF++QTV+R
Sbjct 792 EDHSGGHSMTDIFIHQLIETIEFSLGIISNTASYLRLWALSLSHQQLSAVFFNQTVLR-- 849
Query 169 TLSEDTAVVAA--ALFLVFSLFACI 191
TLS ++ VV +LF +LFA I
Sbjct 850 TLSGESGVVGTTISLFFTSTLFAVI 874
> tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=1015
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 46/97 (47%), Positives = 64/97 (65%), Gaps = 7/97 (7%)
Query 102 QQQQQQQQQQEEHEGAGD-------LFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQ 154
++ +++ + ++ +GA D +FI QMIETIEFVLGT+SNTASYLRLWALSLAHQQ
Sbjct 865 RRTRKRTEVEDAEDGADDGAGEISEVFIHQMIETIEFVLGTISNTASYLRLWALSLAHQQ 924
Query 155 LSLVFYSQTVVRALTLSEDTAVVAAALFLVFSLFACI 191
L+LVF+ +T+ AL + +F +FA I
Sbjct 925 LALVFFEKTIGLALQPGTGGVAMTIKFVFLFPIFALI 961
> pfa:PF08_0113 vacuolar proton translocating ATPase subunit A,
putative; K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1053
Score = 84.0 bits (206), Expect = 3e-16, Method: Composition-based stats.
Identities = 41/83 (49%), Positives = 58/83 (69%), Gaps = 1/83 (1%)
Query 109 QQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRAL 168
++ E +++I Q+IETIEF+LG +SNTASYLRLWALSLAHQQLS VF+ QT++ +L
Sbjct 922 EENHHEENISEIWIEQLIETIEFILGLISNTASYLRLWALSLAHQQLSFVFFEQTILNSL 981
Query 169 TLSEDTAVVAAALFLVFSLFACI 191
+ +V+ L L LF+ +
Sbjct 982 KRNSFMSVL-INLILFSQLFSIL 1003
> tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=936
Score = 83.6 bits (205), Expect = 4e-16, Method: Composition-based stats.
Identities = 44/86 (51%), Positives = 53/86 (61%), Gaps = 7/86 (8%)
Query 98 DAPPQQQQQQQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSL 157
+A P +Q + E LFI Q IETIEF LGT+SNTASYLRLWALSL+HQQLSL
Sbjct 795 EASPAEQPHSLKLSE-------LFIHQFIETIEFTLGTISNTASYLRLWALSLSHQQLSL 847
Query 158 VFYSQTVVRALTLSEDTAVVAAALFL 183
V + Q + L S V+ LF+
Sbjct 848 VLFKQLIFNCLDNSTSLLVMIFGLFI 873
> mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lysosomal
V0 subunit A4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=833
Score = 80.5 bits (197), Expect = 4e-15, Method: Composition-based stats.
Identities = 45/121 (37%), Positives = 69/121 (57%), Gaps = 7/121 (5%)
Query 71 KQQQRAAAPGHDALDAAENGNAAAAAADAPPQQQQQQQQQQEEHEGAGDLFIFQMIETIE 130
K Q ++ DA++ +G+++ A A + EE GD+F+ Q I TIE
Sbjct 668 KSQLQSFTIHEDAVEGDHSGHSSKKTAGA-----HGMKDGHEEEFNFGDIFVHQAIHTIE 722
Query 131 FVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVAAALFLVFSLFAC 190
+ LG +SNTASYLRLWALSLAH +LS V ++ + L L +V +F++F++FA
Sbjct 723 YCLGCISNTASYLRLWALSLAHAELSEVLWTMVMSIGLRLQGWAGLV--GVFIIFAVFAV 780
Query 191 I 191
+
Sbjct 781 L 781
> ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 76.3 bits (186), Expect = 6e-14, Method: Composition-based stats.
Identities = 38/76 (50%), Positives = 52/76 (68%), Gaps = 1/76 (1%)
Query 111 QEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTL 170
EE E + ++F+ Q+I TIEFVLG VSNTASYLRLWALSLAH +LS VFY + ++ A
Sbjct 697 HEEFEFS-EIFVHQLIHTIEFVLGAVSNTASYLRLWALSLAHSELSSVFYEKVLLLAWGY 755
Query 171 SEDTAVVAAALFLVFS 186
+ ++ L +F+
Sbjct 756 NNPLILIVGVLVFIFA 771
> xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysosomal
V0 subunit a4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=846
Score = 75.9 bits (185), Expect = 7e-14, Method: Composition-based stats.
Identities = 35/74 (47%), Positives = 53/74 (71%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
GD+F+ Q I TIE+ LG +SNTASYLRLWALSLAH QLS V ++ + + L+++ ++
Sbjct 723 GDIFVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWTMVMHQGLSIATWGGLI 782
Query 178 AAALFLVFSLFACI 191
+F++F+ FA +
Sbjct 783 --GVFIIFAAFAVL 794
> cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=899
Score = 75.9 bits (185), Expect = 8e-14, Method: Composition-based stats.
Identities = 47/120 (39%), Positives = 61/120 (50%), Gaps = 5/120 (4%)
Query 71 KQQQRAAAPGHDALDAAENGNAAAAAADAPPQQQQQQQQQQEEHEGA----GDLFIFQMI 126
K+++ GH L + N A P+Q + H GD+ ++Q I
Sbjct 721 KEEKERREGGHRQLSVRADINQDDAEVVHAPEQTPKPSGHGHGHGDGPLEMGDVMVYQAI 780
Query 127 ETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVAA-ALFLVF 185
TIEFVLG VS+TASYLRLWALSLAH QLS V ++ A L T +A LF +F
Sbjct 781 HTIEFVLGCVSHTASYLRLWALSLAHAQLSDVLWTMVFRNAFVLDGYTGAIATYILFFIF 840
> ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=817
Score = 75.5 bits (184), Expect = 9e-14, Method: Composition-based stats.
Identities = 34/58 (58%), Positives = 44/58 (75%), Gaps = 0/58 (0%)
Query 110 QQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRA 167
EE ++F+ Q+I +IEFVLG+VSNTASYLRLWALSLAH +LS VFY + ++ A
Sbjct 696 HHEEEFNFSEIFVHQLIHSIEFVLGSVSNTASYLRLWALSLAHSELSTVFYEKVLLLA 753
> ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 75.1 bits (183), Expect = 1e-13, Method: Composition-based stats.
Identities = 40/94 (42%), Positives = 54/94 (57%), Gaps = 9/94 (9%)
Query 102 QQQQQQQQQQEEHEGAG---------DLFIFQMIETIEFVLGTVSNTASYLRLWALSLAH 152
Q + + Q E G G ++F+ Q+I TIEFVLG VSNTASYLRLWALSLAH
Sbjct 678 QLDETDESLQVETNGGGHGHEEFEFSEIFVHQLIHTIEFVLGAVSNTASYLRLWALSLAH 737
Query 153 QQLSLVFYSQTVVRALTLSEDTAVVAAALFLVFS 186
+LS VFY + ++ A + + L +F+
Sbjct 738 SELSSVFYEKVLLMAWGFNNVFIWIVGILVFIFA 771
> dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=849
Score = 75.1 bits (183), Expect = 1e-13, Method: Composition-based stats.
Identities = 41/73 (56%), Positives = 49/73 (67%), Gaps = 2/73 (2%)
Query 119 DLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVA 178
DLF+ Q I TIE+ LG +SNTASYLRLWALSLAH QLS V + +V + L DT+V
Sbjct 724 DLFLNQSIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWE--MVMRVALHVDTSVGI 781
Query 179 AALFLVFSLFACI 191
L VF LFA +
Sbjct 782 VLLVPVFGLFAVL 794
> dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-like;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 74.7 bits (182), Expect = 2e-13, Method: Composition-based stats.
Identities = 39/74 (52%), Positives = 50/74 (67%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
GD+F+ Q I TIE+ LG +SNTASYLRLWALSLAH QLS V ++ + L +S V+
Sbjct 713 GDVFLHQAIHTIEYSLGCISNTASYLRLWALSLAHAQLSEVLWAMVMRLGLRISSRLGVI 772
Query 178 AAALFLVFSLFACI 191
L VFS+FA +
Sbjct 773 --FLVPVFSVFAVL 784
> cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=865
Score = 74.7 bits (182), Expect = 2e-13, Method: Composition-based stats.
Identities = 38/77 (49%), Positives = 52/77 (67%), Gaps = 3/77 (3%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLS--EDTA 175
GD+ + Q I TIEFVLG VS+TASYLRLWALSLAH QLS V ++ + +LT+ +A
Sbjct 739 GDIMVHQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSDVLWTMVLRMSLTMGGWGGSA 798
Query 176 VVAAALFLVFSLFA-CI 191
+ + +FS+ + CI
Sbjct 799 AITILFYFIFSILSVCI 815
> cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 transmembrane
regions near C-terminus ; K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=920
Score = 73.9 bits (180), Expect = 3e-13, Method: Composition-based stats.
Identities = 32/63 (50%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 106 QQQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVV 165
+ ++ H ++FI Q+IET+EF++G++SNTASYLRLWALSLAH L+LV T++
Sbjct 780 HEVEESSGHSDPTEIFIHQLIETVEFLIGSISNTASYLRLWALSLAHNMLALVALQFTIM 839
Query 166 RAL 168
+AL
Sbjct 840 KAL 842
> hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,
RDRTA2, RTA1C, RTADR, STV1, VPH1, VPP2; ATPase, H+ transporting,
lysosomal V0 subunit a4 (EC:3.6.3.14); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=840
Score = 73.6 bits (179), Expect = 4e-13, Method: Composition-based stats.
Identities = 36/74 (48%), Positives = 50/74 (67%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
GD+F+ Q I TIE+ LG +SNTASYLRLWALSLAH QLS V ++ + L +V
Sbjct 717 GDVFVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWTMVMNSGLQTRGWGGIV 776
Query 178 AAALFLVFSLFACI 191
+F++F++FA +
Sbjct 777 --GVFIIFAVFAVL 788
> xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysosomal
V0 subunit a1 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=831
Score = 73.2 bits (178), Expect = 5e-13, Method: Composition-based stats.
Identities = 43/91 (47%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 102 QQQQQQQQQQEEHE-GAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFY 160
Q Q EE E GD + Q I TIE+ LG +SNTASYLRLWALSLAH QLS V +
Sbjct 691 QHDQLSMHSDEEEEFDFGDTVVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLW 750
Query 161 SQTVVRALTLSEDTAVVAAALFLVFSLFACI 191
T+V + L+ + AL +FS FA +
Sbjct 751 --TMVMHIGLNIRSLGGGIALVFIFSAFATL 779
> mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,
TIRC7, Vph1, oc; T-cell, immune regulator 1, ATPase, H+ transporting,
lysosomal V0 protein A3 (EC:3.6.3.6); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 65/123 (52%), Gaps = 1/123 (0%)
Query 70 DKQQQRAAAPGHDALDAAENGNAAAAAADAPPQQQQQQQQQQEEHEGA-GDLFIFQMIET 128
+ Q++ A D + +A+ P +++ EE E ++F+ Q I T
Sbjct 665 NTQRRPAGQQDEDTDKLLASPDASTLENSWSPDEEKAGSPGDEETEFVPSEIFMHQAIHT 724
Query 129 IEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVAAALFLVFSLF 188
IEF LG +SNTASYLRLWALSLAH QLS V ++ + L + + V A L VF+ F
Sbjct 725 IEFCLGCISNTASYLRLWALSLAHAQLSEVLWAMVMRIGLGMGREIGVAAVVLVPVFAAF 784
Query 189 ACI 191
A +
Sbjct 785 AVL 787
> dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0
subunit a1
Length=803
Score = 72.8 bits (177), Expect = 7e-13, Method: Composition-based stats.
Identities = 37/74 (50%), Positives = 50/74 (67%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
GD+F++Q I TIE+ LG +SNTASYLRLWALSLAH +LS V + + L LS +
Sbjct 682 GDVFVYQAIHTIEYCLGCISNTASYLRLWALSLAHAELSEVLWRMVLQAGLKLS--FGLG 739
Query 178 AAALFLVFSLFACI 191
+ L L+F+ FA +
Sbjct 740 SLMLALLFAAFAVL 753
> hsa:10312 TCIRG1, ATP6N1C, ATP6V0A3, Atp6i, OC-116kDa, OC116,
OPTB1, Stv1, TIRC7, Vph1, a3; T-cell, immune regulator 1,
ATPase, H+ transporting, lysosomal V0 subunit A3 (EC:3.6.3.6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=830
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 9/124 (7%)
Query 69 ADKQQQRAAAPGHDALDAAENGNAAAAAADAPPQQQQQQQQQQEEHE-GAGDLFIFQMIE 127
AD+Q++ A D DA+ NG ++ +++ +EE E ++ + Q I
Sbjct 668 ADRQEENKAG-LLDLPDASVNGWSSD-------EEKAGGLDDEEEAELVPSEVLMHQAIH 719
Query 128 TIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVAAALFLVFSL 187
TIEF LG VSNTASYLRLWALSLAH QLS V ++ + L L + V A L +F+
Sbjct 720 TIEFCLGCVSNTASYLRLWALSLAHAQLSEVLWAMVMRIGLGLGREVGVAAVVLVPIFAA 779
Query 188 FACI 191
FA +
Sbjct 780 FAVM 783
> sce:YOR270C VPH1; Subunit a of vacuolar-ATPase V0 domain, one
of two isoforms (Vph1p and Stv1p); Vph1p is located in V-ATPase
complexes of the vacuole while Stv1p is located in V-ATPase
complexes of the Golgi and endosomes (EC:3.6.3.14); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=840
Score = 71.6 bits (174), Expect = 1e-12, Method: Composition-based stats.
Identities = 48/121 (39%), Positives = 63/121 (52%), Gaps = 14/121 (11%)
Query 81 HDALDAAENGNAAAAAADAPPQQ---QQQQQQQQEEHEGAG-------DLFIFQMIETIE 130
H+ L + E A A++ D QQ +EE G+G D+ I Q+I TIE
Sbjct 665 HEPLPSTE---ADASSEDLEAQQLISAMDADDAEEEEVGSGSHGEDFGDIMIHQVIHTIE 721
Query 131 FVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAV-VAAALFLVFSLFA 189
F L VS+TASYLRLWALSLAH QLS V ++ T+ A V + ALF ++
Sbjct 722 FCLNCVSHTASYLRLWALSLAHAQLSSVLWTMTIQIAFGFRGFVGVFMTVALFAMWFALT 781
Query 190 C 190
C
Sbjct 782 C 782
> cel:C26H9A.1 vha-7; Vacuolar H ATPase family member (vha-7);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1213
Score = 71.6 bits (174), Expect = 2e-12, Method: Composition-based stats.
Identities = 39/86 (45%), Positives = 52/86 (60%), Gaps = 10/86 (11%)
Query 111 QEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTL 170
++ H D+F+ Q I TIEFVLG VS+TASYLRLWALSLAH QLS V + +++ +
Sbjct 1056 EDIHHSLSDIFVHQAIHTIEFVLGCVSHTASYLRLWALSLAHAQLSEVMWHMVLIQGIHT 1115
Query 171 -----SEDTA-----VVAAALFLVFS 186
+E A VVA F +F+
Sbjct 1116 VDHIENETIAMCLKPVVACVAFFIFA 1141
> dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator
1, ATPase, H+ transporting, lysosomal V0 subunit A3; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 38/73 (52%), Positives = 51/73 (69%), Gaps = 2/73 (2%)
Query 119 DLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVA 178
++F+ Q I TIE+ LG +SNTASYLRLWALSLAH QL+ V + +V ++LS V +
Sbjct 703 EVFMQQAIHTIEYCLGCISNTASYLRLWALSLAHAQLAEVLW--VMVMRISLSWQGYVGS 760
Query 179 AALFLVFSLFACI 191
L +VFSLFA +
Sbjct 761 VVLSVVFSLFATL 773
> mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1,
Vpp-1, Vpp1; ATPase, H+ transporting, lysosomal V0 subunit
A1 (EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 38/90 (42%), Positives = 53/90 (58%), Gaps = 2/90 (2%)
Query 102 QQQQQQQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYS 161
+ ++ ++E GD + Q I TIE+ LG +SNTASYLRLWALSLAH QLS V +
Sbjct 699 HSEDAEEPTEDEVFDFGDTMVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLW- 757
Query 162 QTVVRALTLSEDTAVVAAALFLVFSLFACI 191
T+V + L + LF +F+ FA +
Sbjct 758 -TMVIHIGLHVRSLAGGLGLFFIFAAFATL 786
> sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=890
Score = 70.5 bits (171), Expect = 4e-12, Method: Composition-based stats.
Identities = 47/117 (40%), Positives = 67/117 (57%), Gaps = 9/117 (7%)
Query 73 QQRAAAPGHDALDAAENGNAAAAAADAPPQQQQQQQQQQEEHEGAGDLFIFQMIETIEFV 132
QQR +A G + ++ A+ AD+ + +Q GD+ I Q+I TIEF
Sbjct 731 QQRHSAEGFQGMIISD----VASVADSINESVGGGEQGPFNF---GDVMIHQVIHTIEFC 783
Query 133 LGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVVAAALFLVFSLFA 189
L +S+TASYLRLWALSLAH QLS V + T+ A + S+++ A + +VF LFA
Sbjct 784 LNCISHTASYLRLWALSLAHAQLSSVLWDMTISNAFS-SKNSGSPLAVMKVVF-LFA 838
> dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1b; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 70.1 bits (170), Expect = 5e-12, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
GD+ + Q I TIE+ LG +SNTASYLRLWALSLAH QLS V + +V L LS +
Sbjct 716 GDMAVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLWG--MVMRLGLSSRSGGG 773
Query 178 AAALFLVFSLFACI 191
L ++FS FA +
Sbjct 774 FFGLSIIFSAFATL 787
> dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1a (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 69.7 bits (169), Expect = 6e-12, Method: Composition-based stats.
Identities = 42/91 (46%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 102 QQQQQQQQQQEEHE-GAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFY 160
Q Q Q +EE E D + Q I TIE+ LG +SNTASYLRLWALSLAH QLS V +
Sbjct 694 QHDQLSQNTEEEPEFNFADEAVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQLSEVLW 753
Query 161 SQTVVRALTLSEDTAVVAAALFLVFSLFACI 191
S +V + LS + L ++F FA +
Sbjct 754 S--MVMHMGLSSRSFGGFIFLSIIFCFFAVL 782
> dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 69.3 bits (168), Expect = 7e-12, Method: Composition-based stats.
Identities = 40/103 (38%), Positives = 64/103 (62%), Gaps = 4/103 (3%)
Query 87 AENGNAAAAAADAPPQQQQQQQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLW 146
AENG+ + D + ++++ + ++F+ Q I TIE+ LG +SNTASYLRLW
Sbjct 673 AENGSINSQQGDVDARGGGGGEEEEFD---TANVFMHQAIHTIEYCLGCISNTASYLRLW 729
Query 147 ALSLAHQQLSLVFYSQTVVRAL-TLSEDTAVVAAALFLVFSLF 188
ALSLAH QLS V ++ + ++ LS +V+AA +F+ F++
Sbjct 730 ALSLAHAQLSEVLWTMVMRQSFGQLSYVGSVMAALVFVGFAVL 772
> dre:100147923 T-cell immune regulator 1-like; K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=793
Score = 69.3 bits (168), Expect = 8e-12, Method: Composition-based stats.
Identities = 40/103 (38%), Positives = 64/103 (62%), Gaps = 5/103 (4%)
Query 87 AENGNAAAAAADAPPQQQQQQQQQQEEHEGAGDLFIFQMIETIEFVLGTVSNTASYLRLW 146
AENG+ + D + ++++ + ++F+ Q I TIE+ LG +SNTASYLRLW
Sbjct 673 AENGSINSQQGDVDARGGGGEEEEFD----TANVFMHQAIHTIEYCLGCISNTASYLRLW 728
Query 147 ALSLAHQQLSLVFYSQTVVRAL-TLSEDTAVVAAALFLVFSLF 188
ALSLAH QLS V ++ + ++ LS +V+AA +F+ F++
Sbjct 729 ALSLAHAQLSEVLWTMVMRQSFGQLSYVGSVMAALVFVGFAVL 771
> cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=873
Score = 67.0 bits (162), Expect = 4e-11, Method: Composition-based stats.
Identities = 32/51 (62%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 115 EGAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVV 165
E GD+ + Q I TIE+VLG VS+TASYLRLWALSLAH QLS V + V
Sbjct 746 ESFGDIMVHQAIHTIEYVLGCVSHTASYLRLWALSLAHAQLSEVLWHMVFV 796
> hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1,
Vph1, a1; ATPase, H+ transporting, lysosomal V0 subunit a1
(EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 66.2 bits (160), Expect = 6e-11, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 45/75 (60%), Gaps = 2/75 (2%)
Query 98 DAPPQQQQQQQQQQEEHE--GAGDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQL 155
DA Q Q E+ + GD + Q I TIE+ LG +SNTASYLRLWALSLAH QL
Sbjct 693 DAEIIQHDQLSTHSEDADEFDFGDTMVHQAIHTIEYCLGCISNTASYLRLWALSLAHAQL 752
Query 156 SLVFYSQTVVRALTL 170
S V ++ + L++
Sbjct 753 SEVLWTMVIHIGLSV 767
> hsa:23545 ATP6V0A2, A2, ARCL, ATP6A2, ATP6N1D, J6B7, RTF, STV1,
TJ6, TJ6M, TJ6S, VPH1, WSS; ATPase, H+ transporting, lysosomal
V0 subunit a2 (EC:3.6.3.6); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=856
Score = 66.2 bits (160), Expect = 6e-11, Method: Composition-based stats.
Identities = 35/74 (47%), Positives = 49/74 (66%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
G++ + Q+I +IE+ LG +SNTASYLRLWALSLAH QLS V ++ ++ + L DT
Sbjct 729 GEILMTQVIHSIEYCLGCISNTASYLRLWALSLAHAQLSDVLWA--MLMRVGLRVDTTYG 786
Query 178 AAALFLVFSLFACI 191
L V +LFA +
Sbjct 787 VLLLLPVIALFAVL 800
> mmu:21871 Atp6v0a2, 8430408C20Rik, AI385560, ATP6a2, AW489264,
Atp6n1d, Atp6n2, C76904, MGC124341, MGC124342, Stv1, TJ6M,
TJ6s, Tj6; ATPase, H+ transporting, lysosomal V0 subunit A2
(EC:3.6.3.6); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=856
Score = 64.3 bits (155), Expect = 3e-10, Method: Composition-based stats.
Identities = 34/74 (45%), Positives = 47/74 (63%), Gaps = 2/74 (2%)
Query 118 GDLFIFQMIETIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTAVV 177
G++ + Q I +IE+ LG +SNTASYLRLWALSLAH QLS V ++ ++ + L DT
Sbjct 729 GEILMTQAIHSIEYCLGCISNTASYLRLWALSLAHAQLSDVLWA--MLMRVGLRVDTTYG 786
Query 178 AAALFLVFSLFACI 191
L V + FA +
Sbjct 787 VLLLLPVMAFFAVL 800
> hsa:57650 CIP2A, FLJ12850, MGC163436, p90; KIAA1524
Length=905
Score = 32.0 bits (71), Expect = 1.2, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 128 TIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTA 175
+++ + T+SN S+ R LAH L++V ++ +++ +LTL+E+
Sbjct 175 SVQTHIKTLSNVKSFYRTLITLLAHSSLTVVVFALSILSSLTLNEEVG 222
> mmu:224171 C330027C09Rik, AA408511, AU018569, Cip2a, Kiaa1524;
RIKEN cDNA C330027C09 gene
Length=907
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 128 TIEFVLGTVSNTASYLRLWALSLAHQQLSLVFYSQTVVRALTLSEDTA 175
+++ + T+SN S+ R LAH L++V ++ +++ +LTL+E+
Sbjct 177 SVQTQIKTLSNVKSFYRTLISFLAHSSLTVVVFALSILSSLTLNEEVG 224
Lambda K H
0.318 0.128 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5493959020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40