bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8011_orf1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
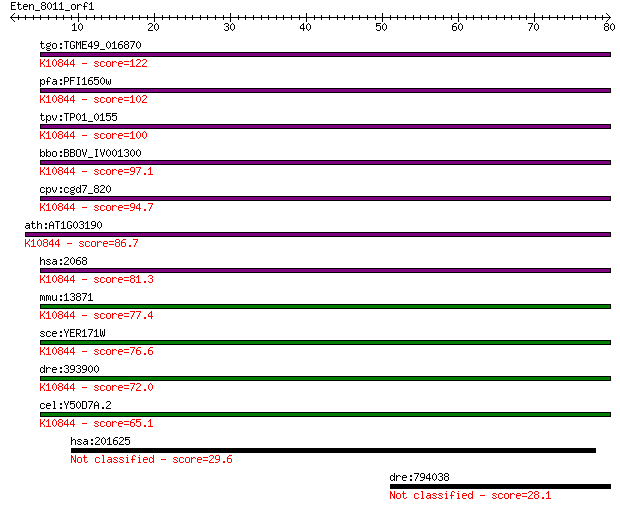
tgo:TGME49_016870 excision repair protein rad15, putative ; K1... 122 2e-28
pfa:PFI1650w DNA excision-repair helicase, putative; K10844 DN... 102 3e-22
tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision rep... 100 8e-22
bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10... 97.1 1e-20
cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision ... 94.7 6e-20
ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP b... 86.7 2e-17
hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219, T... 81.3 6e-16
mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD; ex... 77.4 1e-14
sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nuc... 76.6 2e-14
dre:393900 ercc2, MGC56365, zgc:56365; excision repair cross-c... 72.0 4e-13
cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair ... 65.1 5e-11
hsa:201625 DNAH12, DHC3, DLP12, DNAH12L, DNAH7L, DNAHC12, DNAH... 29.6 2.5
dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting ... 28.1 8.2
> tgo:TGME49_016870 excision repair protein rad15, putative ;
K10844 DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1065
Score = 122 bits (307), Expect = 2e-28, Method: Composition-based stats.
Identities = 57/75 (76%), Positives = 65/75 (86%), Gaps = 0/75 (0%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
LQ+T++E PLTLVADFCTLV TY +GFI+I DPYPEA GL+DPLLQL CLDASLAMQ
Sbjct 657 LQITEVESYAPLTLVADFCTLVATYCEGFILICDPYPEAVGLYDPLLQLSCLDASLAMQP 716
Query 65 VLSRFKSVILTSGTI 79
VL RF+S+ILTSGTI
Sbjct 717 VLKRFQSLILTSGTI 731
> pfa:PFI1650w DNA excision-repair helicase, putative; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1056
Score = 102 bits (255), Expect = 3e-22, Method: Composition-based stats.
Identities = 41/75 (54%), Positives = 62/75 (82%), Gaps = 0/75 (0%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
LQ+ ++E L +V +FCTL+G Y+ GFI+I +PYPEA+G++DPL+Q CLD+S+AM+
Sbjct 682 LQIVNIEDYSSLNIVCNFCTLLGNYFKGFIIICEPYPEATGIYDPLIQFACLDSSIAMKT 741
Query 65 VLSRFKSVILTSGTI 79
V++++KS+ILTSGTI
Sbjct 742 VINKYKSIILTSGTI 756
> tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=894
Score = 100 bits (250), Expect = 8e-22, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 61/75 (81%), Gaps = 1/75 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
L++T + L L LV DFCTLVGTY++GFIVIVDP+P+A+ +DPLLQ CLDAS+AM+
Sbjct 524 LKMTAVGDLTSLHLVVDFCTLVGTYYNGFIVIVDPFPKATS-YDPLLQFSCLDASIAMKS 582
Query 65 VLSRFKSVILTSGTI 79
V+ F+SVILTSGTI
Sbjct 583 VIDNFQSVILTSGTI 597
> bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=822
Score = 97.1 bits (240), Expect = 1e-20, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 59/75 (78%), Gaps = 1/75 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
L +T L L + LVADFCTLVGTY GFIVIV+PYP+ S L++P++Q CLDAS+AMQ
Sbjct 450 LSITALGDLSSIQLVADFCTLVGTYTTGFIVIVEPYPQGS-LYEPVIQFSCLDASIAMQP 508
Query 65 VLSRFKSVILTSGTI 79
V+ F+SVILTSGTI
Sbjct 509 VVENFQSVILTSGTI 523
> cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=841
Score = 94.7 bits (234), Expect = 6e-20, Method: Composition-based stats.
Identities = 41/75 (54%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
L++ D ++ L LV F T++G+Y GFIVIVDPYPE SGL+DP++QL CLD+S+AM+
Sbjct 489 LRIVDSDQYSSLELVCTFFTILGSYSKGFIVIVDPYPEVSGLYDPVIQLSCLDSSIAMRP 548
Query 65 VLSRFKSVILTSGTI 79
+L R++S++LTSGT+
Sbjct 549 ILKRYQSIVLTSGTL 563
> ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP
binding / ATP-dependent DNA helicase/ ATP-dependent helicase/
DNA binding / hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=758
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 41/78 (52%), Positives = 56/78 (71%), Gaps = 1/78 (1%)
Query 3 LALQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPE-ASGLHDPLLQLCCLDASLA 61
L L++TD + LP+ V DF TLVGTY GF +I++PY E + DP+LQL C DASLA
Sbjct 384 LTLEITDTDEFLPIQTVCDFATLVGTYARGFSIIIEPYDERMPHIPDPILQLSCHDASLA 443
Query 62 MQQVLSRFKSVILTSGTI 79
++ V RF+SV++TSGT+
Sbjct 444 IKPVFDRFQSVVITSGTL 461
> hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219,
TTD, XPD; excision repair cross-complementing rodent repair
deficiency, complementation group 2 (EC:3.6.4.12); K10844 DNA
excision repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 81.3 bits (199), Expect = 6e-16, Method: Composition-based stats.
Identities = 37/76 (48%), Positives = 56/76 (73%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEAS-GLHDPLLQLCCLDASLAMQ 63
L++TDL PLTL+A+F TLV TY GF +I++P+ + + + +P+L C+DASLA++
Sbjct 386 LEITDLADFSPLTLLANFATLVSTYAKGFTIIIEPFDDRTPTIANPILHFSCMDASLAIK 445
Query 64 QVLSRFKSVILTSGTI 79
V RF+SVI+TSGT+
Sbjct 446 PVFERFQSVIITSGTL 461
> mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD;
excision repair cross-complementing rodent repair deficiency,
complementation group 2 (EC:3.6.4.12); K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/76 (47%), Positives = 55/76 (72%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEAS-GLHDPLLQLCCLDASLAMQ 63
L++ DL PLTL+A+F TLV TY GF +I++P+ + + + +P+L C+DASLA++
Sbjct 386 LEIADLADFSPLTLLANFATLVSTYAKGFTIIIEPFDDRTPTIANPVLHFSCMDASLAIK 445
Query 64 QVLSRFKSVILTSGTI 79
V RF+SVI+TSGT+
Sbjct 446 PVFERFQSVIITSGTL 461
> sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nucleotide
excision repair and transcription; subunit of RNA
polymerase II transcription initiation factor TFIIH; subunit
of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human
XPD protein (EC:3.6.1.-); K10844 DNA excision repair protein
ERCC-2 [EC:3.6.4.12]
Length=778
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/76 (46%), Positives = 55/76 (72%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYP-EASGLHDPLLQLCCLDASLAMQ 63
L+VT++E L +A F TL+ TY +GF++I++PY E + + +P+++ CLDAS+A++
Sbjct 388 LEVTEVEDFTALKDIATFATLISTYEEGFLLIIEPYEIENAAVPNPIMRFTCLDASIAIK 447
Query 64 QVLSRFKSVILTSGTI 79
V RF SVI+TSGTI
Sbjct 448 PVFERFSSVIITSGTI 463
> dre:393900 ercc2, MGC56365, zgc:56365; excision repair cross-complementing
rodent repair deficiency, complementation group
2; K10844 DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=643
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 30/76 (39%), Positives = 53/76 (69%), Gaps = 1/76 (1%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEAS-GLHDPLLQLCCLDASLAMQ 63
L++ D+ P+TL++ F TLV TY GF +I++P+ + + + +P+L C+D S+A++
Sbjct 386 LEIADIADFSPITLISHFATLVSTYSKGFTIIIEPFEDKTPTIANPVLHFSCMDPSIAIK 445
Query 64 QVLSRFKSVILTSGTI 79
V RF++VI+TSGT+
Sbjct 446 PVFGRFQTVIITSGTL 461
> cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=606
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 49/75 (65%), Gaps = 3/75 (4%)
Query 5 LQVTDLERLLPLTLVADFCTLVGTYWDGFIVIVDPYPEASGLHDPLLQLCCLDASLAMQQ 64
L++TD + L+ V CTLV TY GF VIV+P G ++ L C DAS+A++
Sbjct 235 LEITDNGDVWALSQVTTLCTLVSTYSKGFSVIVEP---QDGSQLAVITLSCHDASIAIRP 291
Query 65 VLSRFKSVILTSGTI 79
V+SRF+SVI+TSGT+
Sbjct 292 VMSRFQSVIITSGTL 306
> hsa:201625 DNAH12, DHC3, DLP12, DNAH12L, DNAH7L, DNAHC12, DNAHC3,
DNHD2, FLJ40427, FLJ44290, HDHC3, HL-19, HL19; dynein,
axonemal, heavy chain 12
Length=3092
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 39/93 (41%), Gaps = 30/93 (32%)
Query 9 DLERLLPLTLVADFCTLV--------------------GTYWDGFIVI----VDPYPEAS 44
D +R L LT++ADF L GTY D I +PE
Sbjct 2716 DWDRRLLLTMLADFYNLYIVENPHYKFSPSGNYFAPPKGTYEDYIEFIKKLPFTQHPEIF 2775
Query 45 GLHDPLLQLCCLDASLAMQQVLSRFKSVILTSG 77
GLH+ + D S +QQ + F+S++LT G
Sbjct 2776 GLHENV------DISKDLQQTKTLFESLLLTQG 2802
> dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting
protein C-terminal helicase 1
Length=1217
Score = 28.1 bits (61), Expect = 8.2, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 51 LQLCCLDASLAMQQVLSRFKSVILTSGTI 79
L CL+ ++A + S +S++LTSGT+
Sbjct 618 LSFWCLNPAVAFSDLSSTVRSIVLTSGTL 646
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2063098576
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40