bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8006_orf1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
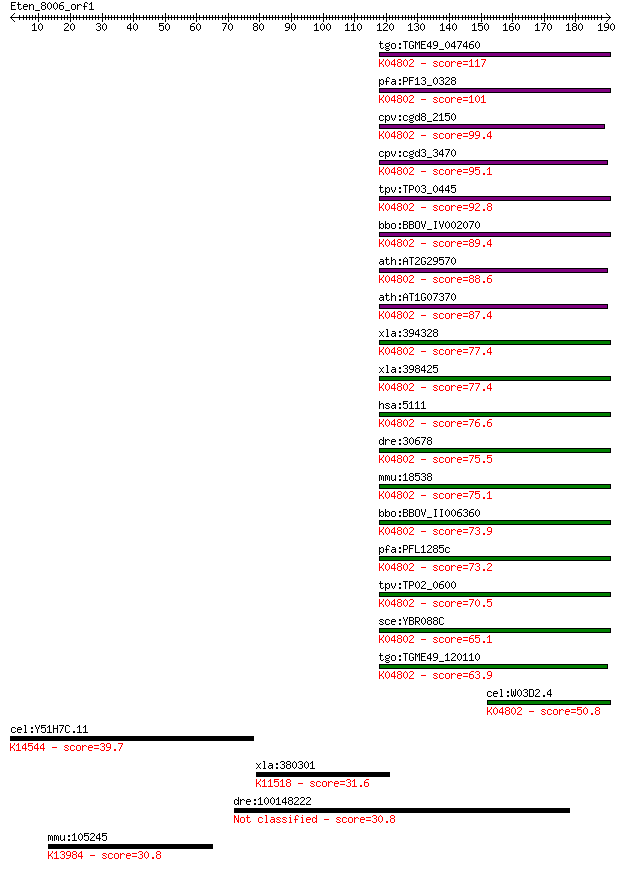
tgo:TGME49_047460 proliferating cell nuclear antigen 1 ; K0480... 117 3e-26
pfa:PF13_0328 proliferating cell nuclear antigen; K04802 proli... 101 1e-21
cpv:cgd8_2150 proliferating cell nuclear antigen PCNA ; K04802... 99.4 7e-21
cpv:cgd3_3470 proliferating cell nuclear antigen ; K04802 prol... 95.1 1e-19
tpv:TP03_0445 proliferating cell nuclear antigen; K04802 proli... 92.8 6e-19
bbo:BBOV_IV002070 21.m02871; proliferating cell nuclear antige... 89.4 6e-18
ath:AT2G29570 PCNA2; PCNA2 (PROLIFERATING CELL NUCLEAR ANTIGEN... 88.6 1e-17
ath:AT1G07370 PCNA1; PCNA1 (PROLIFERATING CELLULAR NUCLEAR ANT... 87.4 3e-17
xla:394328 pcna, pcna-A; proliferating cell nuclear antigen; K... 77.4 2e-14
xla:398425 MGC53867, PCNA; similar to proliferating cell nucle... 77.4 3e-14
hsa:5111 PCNA, MGC8367; proliferating cell nuclear antigen; K0... 76.6 5e-14
dre:30678 pcna, cb16, fb36g03, wu:fa28e03, wu:fb36g03; prolife... 75.5 1e-13
mmu:18538 Pcna; proliferating cell nuclear antigen; K04802 pro... 75.1 1e-13
bbo:BBOV_II006360 18.m06523; Proliferating cell nuclear antige... 73.9 3e-13
pfa:PFL1285c PCNA2; proliferating cell nuclear antigen 2; K048... 73.2 6e-13
tpv:TP02_0600 proliferating cell nuclear antigen; K04802 proli... 70.5 3e-12
sce:YBR088C POL30; PCNA; K04802 proliferating cell nuclear ant... 65.1 1e-10
tgo:TGME49_120110 proliferating cell nuclear antigen, putative... 63.9 3e-10
cel:W03D2.4 pcn-1; PCNA (Proliferating Cell Nuclear Antigen) h... 50.8 2e-06
cel:Y51H7C.11 nol-6; NucleOLar protein family member (nol-6); ... 39.7 0.006
xla:380301 tomm40, MGC53384; translocase of outer mitochondria... 31.6 1.7
dre:100148222 polyprotein-like 30.8 2.7
mmu:105245 Txndc5, AL022641, ERp46, PC-TRP; thioredoxin domain... 30.8 2.9
> tgo:TGME49_047460 proliferating cell nuclear antigen 1 ; K04802
proliferating cell nuclear antigen
Length=317
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 51/73 (69%), Positives = 68/73 (93%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLEA+LQ+A VL+R+FE++ +V++VNLDCDE+GL+LQAMD SHVALV+LKL+DVGF+H+
Sbjct 1 MLEAKLQHASVLRRLFESIKDMVSDVNLDCDETGLRLQAMDSSHVALVALKLDDVGFVHF 60
Query 178 RCDRPRSLGLNMS 190
RCDR RSLGLN++
Sbjct 61 RCDRERSLGLNLA 73
> pfa:PF13_0328 proliferating cell nuclear antigen; K04802 proliferating
cell nuclear antigen
Length=274
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/73 (63%), Positives = 59/73 (80%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLEA+L NA +LK++FE + LVN+ N+D DESGLKLQA+D +HV+LVSL L D GF HY
Sbjct 1 MLEAKLNNASILKKLFECIKDLVNDANVDADESGLKLQALDGNHVSLVSLHLLDSGFSHY 60
Query 178 RCDRPRSLGLNMS 190
RCDR R LG+N++
Sbjct 61 RCDRERVLGVNIA 73
> cpv:cgd8_2150 proliferating cell nuclear antigen PCNA ; K04802
proliferating cell nuclear antigen
Length=262
Score = 99.4 bits (246), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 43/71 (60%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL N +L++VFEA+T LV++VNL+C+ESG+ +QAMD SHV+LV L L+D F Y
Sbjct 1 MFEARLSNGGILRKVFEAITNLVSDVNLECNESGVTIQAMDNSHVSLVGLYLKDTAFERY 60
Query 178 RCDRPRSLGLN 188
RCD+ R+LGLN
Sbjct 61 RCDKNRTLGLN 71
> cpv:cgd3_3470 proliferating cell nuclear antigen ; K04802 proliferating
cell nuclear antigen
Length=261
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/72 (59%), Positives = 55/72 (76%), Gaps = 0/72 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARLQNA++ K++ EA+ L +++N+DCD GL LQAMD SHVALVSL ++ F HY
Sbjct 1 MFEARLQNAMLFKKIAEALKELCHDINIDCDSDGLHLQAMDSSHVALVSLNIQPDAFEHY 60
Query 178 RCDRPRSLGLNM 189
RCDRP LGL+M
Sbjct 61 RCDRPVVLGLDM 72
> tpv:TP03_0445 proliferating cell nuclear antigen; K04802 proliferating
cell nuclear antigen
Length=274
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 58/73 (79%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLE +L NAVVL+R+F+ + L+ + N+D D +G+ LQA+D +H+ALV LKL + GF+ Y
Sbjct 1 MLELKLNNAVVLRRIFDCIRDLITDGNIDFDATGMTLQALDGNHIALVHLKLHESGFVLY 60
Query 178 RCDRPRSLGLNMS 190
RCDRPR+LG+N++
Sbjct 61 RCDRPRALGININ 73
> bbo:BBOV_IV002070 21.m02871; proliferating cell nuclear antigen
1; K04802 proliferating cell nuclear antigen
Length=277
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 58/73 (79%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLE +L +AVVL+R+F+ M ++++ N+D D +G+ LQA+D +HVALV LKL + GF Y
Sbjct 1 MLELKLNHAVVLRRIFDCMRDIISDGNIDFDATGMSLQALDGNHVALVHLKLHESGFSLY 60
Query 178 RCDRPRSLGLNMS 190
RCDRPR+LG+N++
Sbjct 61 RCDRPRALGINLN 73
> ath:AT2G29570 PCNA2; PCNA2 (PROLIFERATING CELL NUCLEAR ANTIGEN
2); DNA binding / DNA polymerase processivity factor; K04802
proliferating cell nuclear antigen
Length=264
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 42/72 (58%), Positives = 50/72 (69%), Gaps = 0/72 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLE RL +LK+V EA+ LVN+ N DC +G LQAMD SHVALVSL L GF HY
Sbjct 1 MLELRLVQGSLLKKVLEAVKDLVNDANFDCSTTGFSLQAMDSSHVALVSLLLRSEGFEHY 60
Query 178 RCDRPRSLGLNM 189
RCDR S+G+N+
Sbjct 61 RCDRNLSMGMNL 72
> ath:AT1G07370 PCNA1; PCNA1 (PROLIFERATING CELLULAR NUCLEAR ANTIGEN);
DNA binding / DNA polymerase processivity factor/ protein
binding; K04802 proliferating cell nuclear antigen
Length=263
Score = 87.4 bits (215), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/72 (56%), Positives = 50/72 (69%), Gaps = 0/72 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLE RL +LK+V E++ LVN+ N DC +G LQAMD SHVALVSL L GF HY
Sbjct 1 MLELRLVQGSLLKKVLESIKDLVNDANFDCSSTGFSLQAMDSSHVALVSLLLRSEGFEHY 60
Query 178 RCDRPRSLGLNM 189
RCDR S+G+N+
Sbjct 61 RCDRNLSMGMNL 72
> xla:394328 pcna, pcna-A; proliferating cell nuclear antigen;
K04802 proliferating cell nuclear antigen
Length=261
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/73 (50%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL +LK+V EA+ L++E D SG+ LQ+MD SHV+LV L L GF Y
Sbjct 1 MFEARLVQGSILKKVLEALKDLIDEACWDITSSGISLQSMDSSHVSLVQLTLRSDGFDTY 60
Query 178 RCDRPRSLGLNMS 190
RCDR +S+G+ MS
Sbjct 61 RCDRNQSIGVKMS 73
> xla:398425 MGC53867, PCNA; similar to proliferating cell nuclear
antigen; K04802 proliferating cell nuclear antigen
Length=261
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 37/73 (50%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL +LK+V EA+ L++E D SG+ LQ+MD SHV+LV L L GF Y
Sbjct 1 MFEARLVQGSILKKVLEALKDLIDEACWDITSSGISLQSMDSSHVSLVQLTLRSDGFDTY 60
Query 178 RCDRPRSLGLNMS 190
RCDR +S+G+ MS
Sbjct 61 RCDRNQSIGVKMS 73
> hsa:5111 PCNA, MGC8367; proliferating cell nuclear antigen;
K04802 proliferating cell nuclear antigen
Length=261
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL +LK+V EA+ L+NE D SG+ LQ+MD SHV+LV L L GF Y
Sbjct 1 MFEARLVQGSILKKVLEALKDLINEACWDISSSGVNLQSMDSSHVSLVQLTLRSEGFDTY 60
Query 178 RCDRPRSLGLNMS 190
RCDR ++G+N++
Sbjct 61 RCDRNLAMGVNLT 73
> dre:30678 pcna, cb16, fb36g03, wu:fa28e03, wu:fb36g03; proliferating
cell nuclear antigen; K04802 proliferating cell nuclear
antigen
Length=261
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 47/73 (64%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL +LK+V EA+ L+ E D SG+ LQ+MD SHV+LV L L GF Y
Sbjct 1 MFEARLVQGSILKKVLEALKDLITEACWDVSSSGISLQSMDSSHVSLVQLTLRSDGFDSY 60
Query 178 RCDRPRSLGLNMS 190
RCDR ++G+N+S
Sbjct 61 RCDRNLAMGVNLS 73
> mmu:18538 Pcna; proliferating cell nuclear antigen; K04802 proliferating
cell nuclear antigen
Length=261
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 47/73 (64%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M EARL +LK+V EA+ L+NE D G+ LQ+MD SHV+LV L L GF Y
Sbjct 1 MFEARLIQGSILKKVLEALKDLINEACWDVSSGGVNLQSMDSSHVSLVQLTLRSEGFDTY 60
Query 178 RCDRPRSLGLNMS 190
RCDR ++G+N++
Sbjct 61 RCDRNLAMGVNLT 73
> bbo:BBOV_II006360 18.m06523; Proliferating cell nuclear antigen;
K04802 proliferating cell nuclear antigen
Length=264
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 51/73 (69%), Gaps = 1/73 (1%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M E RL + + L+R+FEA+ + N+V++DC E GL +QAMD SH++L+ L L F Y
Sbjct 1 MFECRL-DGMFLRRLFEALREICNDVSIDCSEDGLSMQAMDNSHISLIHLCLAPDFFQLY 59
Query 178 RCDRPRSLGLNMS 190
RCD P +LGLN+S
Sbjct 60 RCDTPCTLGLNIS 72
> pfa:PFL1285c PCNA2; proliferating cell nuclear antigen 2; K04802
proliferating cell nuclear antigen
Length=264
Score = 73.2 bits (178), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 50/73 (68%), Gaps = 1/73 (1%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M E R+ + K++FE + + EVNL+CDE+G+K+Q+MD SHV+LV L + F HY
Sbjct 1 MFECRI-DGQFFKKLFETLKDICTEVNLECDENGIKMQSMDCSHVSLVDLNIVSDFFQHY 59
Query 178 RCDRPRSLGLNMS 190
RCD+ LG++++
Sbjct 60 RCDKNCVLGISIN 72
> tpv:TP02_0600 proliferating cell nuclear antigen; K04802 proliferating
cell nuclear antigen
Length=261
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 51/73 (69%), Gaps = 1/73 (1%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M E +L + + L+R+FE++ + +V++DC + GL +QAMD SH++L+ LKL F Y
Sbjct 1 MFECKL-DGMFLRRLFESLKDICGDVSIDCSDEGLTMQAMDNSHISLIHLKLSPDFFHLY 59
Query 178 RCDRPRSLGLNMS 190
RCD P +LGLN+S
Sbjct 60 RCDLPCNLGLNIS 72
> sce:YBR088C POL30; PCNA; K04802 proliferating cell nuclear antigen
Length=258
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
MLEA+ + A + KR+ + V VN C E G+ QA+D S V LVSL++ F Y
Sbjct 1 MLEAKFEEASLFKRIIDGFKDCVQLVNFQCKEDGIIAQAVDDSRVLLVSLEIGVEAFQEY 60
Query 178 RCDRPRSLGLNMS 190
RCD P +LG++++
Sbjct 61 RCDHPVTLGMDLT 73
> tgo:TGME49_120110 proliferating cell nuclear antigen, putative
; K04802 proliferating cell nuclear antigen
Length=287
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Query 118 MLEARLQNAVVLKRVFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
M E ++ ++LKR+ E + +V +VNL C+ SG+ L +MD SHVA+V ++L F Y
Sbjct 1 MFECTIE-GLLLKRLMECLKEMVTDVNLVCNASGISLDSMDGSHVAVVDVRLAVDLFHKY 59
Query 178 RCDRPRSLGLNM 189
RCDRP LGL++
Sbjct 60 RCDRPVQLGLSV 71
> cel:W03D2.4 pcn-1; PCNA (Proliferating Cell Nuclear Antigen)
homolog family member (pcn-1); K04802 proliferating cell nuclear
antigen
Length=229
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/40 (67%), Positives = 32/40 (80%), Gaps = 2/40 (5%)
Query 152 LKLQAMDQSHVALVSLKLEDVG-FLHYRCDRPRSLGLNMS 190
+ LQAMD SHVALVSLKLE VG F YRCDR +LGL+++
Sbjct 1 MSLQAMDSSHVALVSLKLE-VGLFDTYRCDRTINLGLSLA 39
> cel:Y51H7C.11 nol-6; NucleOLar protein family member (nol-6);
K14544 U3 small nucleolar RNA-associated protein 22
Length=1058
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 38/78 (48%), Gaps = 1/78 (1%)
Query 1 STGKLPELFRIYLFLSTLSSFPANKAPVT-PPPQPPSLPSGLLGYRLSVRLLHISAFLVN 59
S + PE +R +L S K P+T PP + GLL Y+L V L IS FL+
Sbjct 440 SLSEYPEKYRQKTYLIGFRSTSGWKNPLTVGPPAQTNDAKGLLDYKLQVFRLKISIFLLE 499
Query 60 SFRKFHSFFKKKASRASF 77
+ +KF +K+ + F
Sbjct 500 NRKKFRELWKETSELRKF 517
> xla:380301 tomm40, MGC53384; translocase of outer mitochondrial
membrane 40 homolog; K11518 mitochondrial import receptor
subunit TOM40
Length=336
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Query 79 LRASPPGLALPPAAQLAQQPQQQQQQQQQLPK---LSSSHKKMLE 120
L + PPG +PP A L P +++ Q+++LP H+K E
Sbjct 23 LVSVPPGFTMPPVAGLTPTPDKKEPQEERLPNPGTFEECHRKCKE 67
> dre:100148222 polyprotein-like
Length=546
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 14/106 (13%)
Query 72 ASRASFRLRASPPGLALPPAAQLAQQPQQQQQQQQQLPKLSSSHKKMLEARLQNAVVLKR 131
ASR F L ++ P PP LA +P Q +++H ++L + + L
Sbjct 246 ASRPPFTLSSATP--LPPPNNALAMEPPQVS---------NAAHNQILSGADVDLITL-- 292
Query 132 VFEAMTCLVNEVNLDCDESGLKLQAMDQSHVALVSLKLEDVGFLHY 177
+T E +DC E L L+ SH ++SL ++ F Y
Sbjct 293 -LSPITPPTAERQVDCGEYSLTLKQPHSSHSRILSLAEFNIAFSQY 337
> mmu:105245 Txndc5, AL022641, ERp46, PC-TRP; thioredoxin domain
containing 5; K13984 thioredoxin domain-containing protein
5
Length=417
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 22/52 (42%), Gaps = 2/52 (3%)
Query 13 LFLSTLSSFPANKAPVTPPPQPPSLPSGLLGYRLSVRLLHISAFLVNSFRKF 64
L TL+ PA P PP+ P L GL Y LS + N F KF
Sbjct 149 WMLQTLNEEPATPEPEAEPPRAPELKQGL--YELSANNFELHVSQGNHFIKF 198
Lambda K H
0.321 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40