bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7981_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
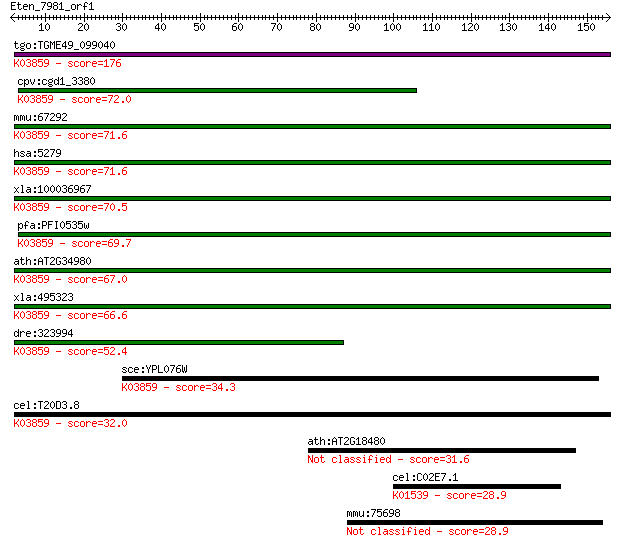
tgo:TGME49_099040 phosphatidylinositol N-acetylglucosaminyltra... 176 2e-44
cpv:cgd1_3380 phosphatidylinositol-glycan-class c, pigC, 8x tr... 72.0 7e-13
mmu:67292 Pigc, 3110030E07Rik, AW212108; phosphatidylinositol ... 71.6 1e-12
hsa:5279 PIGC, GPI2, MGC2049; phosphatidylinositol glycan anch... 71.6 1e-12
xla:100036967 hypothetical protein LOC100036967; K03859 phosph... 70.5 2e-12
pfa:PFI0535w Phosphatidylinositol N-acetylglucosaminyltransfer... 69.7 4e-12
ath:AT2G34980 SETH1; SETH1; phosphatidylinositol N-acetylgluco... 67.0 2e-11
xla:495323 pigc; phosphatidylinositol glycan anchor biosynthes... 66.6 3e-11
dre:323994 pigc, wu:fc16d03, zgc:85894; phosphatidylinositol g... 52.4 6e-07
sce:YPL076W GPI2, GCR4; Gpi2p (EC:2.4.1.198); K03859 phosphati... 34.3 0.17
cel:T20D3.8 hypothetical protein; K03859 phosphatidylinositol ... 32.0 0.72
ath:AT2G18480 mannitol transporter, putative 31.6 1.1
cel:C02E7.1 hypothetical protein; K01539 sodium/potassium-tran... 28.9 6.2
mmu:75698 Fam35a, 3110001K24Rik; family with sequence similari... 28.9 7.3
> tgo:TGME49_099040 phosphatidylinositol N-acetylglucosaminyltransferase
subunit, putative (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=348
Score = 176 bits (447), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 87/154 (56%), Positives = 114/154 (74%), Gaps = 0/154 (0%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLTQ+FS+DTV+ L+ + LLVH L DYSY+YRNP+ +DE L R +S+N AL A+V+LA
Sbjct 185 RTLTQTFSEDTVVCLSVVSLLVHTALTDYSYIYRNPDKVDESLQRAMSINAALLANVVLA 244
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKESSL 121
SRLS+STEVFA L FGIE+F ++P+ARR L W +V TP ++ TAL + E+
Sbjct 245 SRLSSSTEVFAVLIFGIEIFTISPMARRILWQKYPWAFVHVFTPTLVLSTALLLSLEAPA 304
Query 122 GSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
+ L+LF + FITFVGPYWLI SQKYK+EI GP
Sbjct 305 SIVLLFLFSIVFITFVGPYWLISSQKYKHEIKGP 338
> cpv:cgd1_3380 phosphatidylinositol-glycan-class c, pigC, 8x
transmembrane domains (EC:3.5.1.89); K03859 phosphatidylinositol
glycan, class C
Length=274
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 67/105 (63%), Gaps = 4/105 (3%)
Query 3 TLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLAS 62
TLT SFSDDT+I+L +I L+++ +DY+ +Y++ + I+ +++LNL++ S+LLAS
Sbjct 118 TLTASFSDDTIIALCTIAFLLYLLSHDYTIIYKDLKEIEHQNTDVVALNLSMLGSILLAS 177
Query 63 RLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCT--YVLTP 105
RL + +V+ FL F I + + + R LW++ YVLTP
Sbjct 178 RLENNIQVYFFLCFSIHILYFSRMVRHN--LWNNLPKVYLYVLTP 220
> mmu:67292 Pigc, 3110030E07Rik, AW212108; phosphatidylinositol
glycan anchor biosynthesis, class C (EC:2.4.1.198); K03859
phosphatidylinositol glycan, class C
Length=297
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 56/162 (34%), Positives = 86/162 (53%), Gaps = 25/162 (15%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT+S S DT+ ++ LL H+ DY N + L SLN+A+FASV LA
Sbjct 138 KTLTESVSTDTIYAMAVFMLLGHLIFFDYG---ANAAIVSSTL----SLNMAIFASVCLA 190
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTP-AVITLTALFVYKE-- 118
SRL S F + F I++FAL P+ ++ L + TP + + +T LF +
Sbjct 191 SRLPRSLHAFIMVTFAIQIFALWPMLQKKLKAY---------TPRSYVGVTLLFAFSAFG 241
Query 119 -----SSLGSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
S++G++ L+ L+F I+ + PY+LI Q +K I GP
Sbjct 242 GLLSISAVGAI-LFALLLFSISCLCPYYLIHLQLFKENIHGP 282
> hsa:5279 PIGC, GPI2, MGC2049; phosphatidylinositol glycan anchor
biosynthesis, class C (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=297
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 55/161 (34%), Positives = 86/161 (53%), Gaps = 23/161 (14%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT+S S DT+ +++ LL H+ DY N + L SLN+A+FASV LA
Sbjct 138 KTLTESVSTDTIYAMSVFMLLGHLIFFDYG---ANAAIVSSTL----SLNMAIFASVCLA 190
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKE--- 118
SRL S F + F I++FAL P+ ++ L CT + + +T LF +
Sbjct 191 SRLPRSLHAFIMVTFAIQIFALWPMLQKKLK-----ACT---PRSYVGVTLLFAFSAVGG 242
Query 119 ----SSLGSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
S++G++ L+ L+ I+ + P++LIR Q +K I GP
Sbjct 243 LLSISAVGAV-LFALLLMSISCLCPFYLIRLQLFKENIHGP 282
> xla:100036967 hypothetical protein LOC100036967; K03859 phosphatidylinositol
glycan, class C
Length=317
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 58/157 (36%), Positives = 89/157 (56%), Gaps = 15/157 (9%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT+S S DT+ +++ + LL H+ DY N + LS+N+A+FASV LA
Sbjct 158 KTLTESISTDTIYAMSVLMLLGHLVFFDYG---ANAAVVSS----TLSINMAIFASVCLA 210
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSW---GCTYVLTPAVITLTALFVYKE 118
SRL S FA + F I++FAL P +R L + G T++ A++T+ L
Sbjct 211 SRLPRSLHAFAMVTFAIQIFALWPSLQRKLRANTPRTYIGVTFLF--AILTMAGLL--SI 266
Query 119 SSLGSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
S +G++ +L L+ +TF+ PY LIR Q +K+ I GP
Sbjct 267 SGVGALLFFL-LLLSVTFLCPYCLIRLQLFKDNIYGP 302
> pfa:PFI0535w Phosphatidylinositol N-acetylglucosaminyltransferase,
putative (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=292
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 84/153 (54%), Gaps = 3/153 (1%)
Query 3 TLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLAS 62
+LTQ+ SD+TV ++ + LL+H+ + Y ++Y E ID + SL+ + ASV+L S
Sbjct 132 SLTQTHSDNTVYLVSIMLLLIHLMFHKYGFIYEKNENID--IFDATSLSCVVIASVILGS 189
Query 63 RLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKESSLG 122
RL++ +VF+FLF +F +P + + L + YVL P + + +L + + S+
Sbjct 190 RLASIEQVFSFLFVSSILFFYSPFIFQTIALKNINYYNYVLFPFLFVILSLCI-RSISIT 248
Query 123 SMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
+ L FF+ F+ P + ++ K ++ GP
Sbjct 249 LFYVNLIGQFFLLFIVPAFFVKKHNLKTKLEGP 281
> ath:AT2G34980 SETH1; SETH1; phosphatidylinositol N-acetylglucosaminyltransferase/
transferase; K03859 phosphatidylinositol
glycan, class C
Length=303
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 56/156 (35%), Positives = 88/156 (56%), Gaps = 4/156 (2%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYS-YVYRNPETIDEP-LGRLLSLNLALFASVL 59
QTLT+S S D++ ++T LL+H+ L+DYS R P + P L +S+N ++ ASV
Sbjct 138 QTLTRSISSDSIWAVTVSLLLLHLFLHDYSGSTIRAPGALKTPNLTSCISVNASIVASVF 197
Query 60 LASRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKES 119
+ASRL + VFA + F ++VF AP+ Y + ++G + + A++ LT +Y
Sbjct 198 VASRLPSRLHVFAVMLFSLQVFLFAPLV-TYCIKKFNFGLHLLFSFALMGLTLYSIYALH 256
Query 120 SLGSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
L + ++ PYWLIR Q+YK EI+GP
Sbjct 257 RLFFLVFLSLVLLVNVVC-PYWLIRMQEYKFEINGP 291
> xla:495323 pigc; phosphatidylinositol glycan anchor biosynthesis,
class C (EC:2.4.1.198); K03859 phosphatidylinositol glycan,
class C
Length=319
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 54/154 (35%), Positives = 82/154 (53%), Gaps = 9/154 (5%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT+S S DT+ +++ + LL H+ D+ N + L S+N+A+FASV LA
Sbjct 160 KTLTESISTDTIYAMSVLMLLGHLVFFDFG---ANAAVVSSTL----SINMAIFASVCLA 212
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKESSL 121
SRL S FA + F I++FAL P +R L ++ TY+ + + AL S
Sbjct 213 SRLPRSLHAFAMVTFAIQIFALWPSLQRKLR--ANTPRTYIGVTFLFAIFALAGLLSISG 270
Query 122 GSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
L+ L+ + F+ PY LIR Q +K+ I GP
Sbjct 271 VGALLFFLLLLSVAFLCPYCLIRLQLFKDNIHGP 304
> dre:323994 pigc, wu:fc16d03, zgc:85894; phosphatidylinositol
glycan, class C (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=293
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 48/85 (56%), Gaps = 9/85 (10%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT+S S DTV +++++ LL H+ S+ Y P P G L SLN ALF SV LA
Sbjct 136 KTLTESVSTDTVYAMSAVMLLAHL----VSFPYAQPS----PPGSL-SLNAALFGSVCLA 186
Query 62 SRLSTSTEVFAFLFFGIEVFALAPI 86
SRL + F L + VFAL P
Sbjct 187 SRLPGALHTFTMLTCALLVFALWPC 211
> sce:YPL076W GPI2, GCR4; Gpi2p (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=280
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 36/133 (27%), Positives = 61/133 (45%), Gaps = 26/133 (19%)
Query 30 YSYVYRNPETIDEPLGRLLSLNLALFASVLLASRLSTSTEVFAFLFFGIEVFALAPIARR 89
Y +V + ++ D+P LS N+ + +L+SRLST+ +VF FL I++ + P
Sbjct 152 YIFVISSTKSKDKPSN--LSTNILVALVAVLSSRLSTTIDVFCFLLICIQLNIILP---- 205
Query 90 YLLLWSSWGCTY--VLTPAVITLTALFVYKESSLGSMALYLFLMFFITF----VGP---- 139
TY V V ++ + VY ++ +Y+ L+FF + V P
Sbjct 206 ----------TYLSVTNKVVPIISNIIVYSFLNVALGWIYMLLIFFASVFYITVLPKWFI 255
Query 140 YWLIRSQKYKNEI 152
YW I K N++
Sbjct 256 YWKINYHKRDNDL 268
> cel:T20D3.8 hypothetical protein; K03859 phosphatidylinositol
glycan, class C
Length=282
Score = 32.0 bits (71), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 38/154 (24%), Positives = 61/154 (39%), Gaps = 9/154 (5%)
Query 2 QTLTQSFSDDTVISLTSICLLVHIPLNDYSYVYRNPETIDEPLGRLLSLNLALFASVLLA 61
+TLT S S DT+ S + I + +DY + P S++ L +++ L
Sbjct 127 RTLTTSISTDTIYSTSIITAIFSCFFHDYGV---KAPVVSYP----TSVSTGLSSAIFLL 179
Query 62 SRLSTSTEVFAFLFFGIEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKESSL 121
SRL T L + A R ++ + C ++L +++ + SL
Sbjct 180 SRLEGDTPTLLLLVVAFTLHAYGAEFRNR--IFHVYPCLSSTIFCFLSLFSIYCISDFSL 237
Query 122 GSMALYLFLMFFITFVGPYWLIRSQKYKNEISGP 155
+ L FI F+ P LI Q K I GP
Sbjct 238 ELSICFALLHIFILFICPLILILKQTGKCTIHGP 271
> ath:AT2G18480 mannitol transporter, putative
Length=508
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 37/74 (50%), Gaps = 5/74 (6%)
Query 78 IEVFALAPIARRYLLLWSSWGCTYVLTPAVITLTALFVYKES----SLGSMALYLFLMFF 133
I F L + RR LLL S+ G + LT ++LT + + SL ++ Y F+ FF
Sbjct 335 IATFLLDKVGRRKLLLTSTGGMVFALTSLAVSLTMVQRFGRLAWALSLSIVSTYAFVAFF 394
Query 134 ITFVGPY-WLIRSQ 146
+GP W+ S+
Sbjct 395 SIGLGPITWVYSSE 408
> cel:C02E7.1 hypothetical protein; K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1050
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query 100 TYVLTPAVITLTALFVYKESSLGSMALYLFLMFFITFVGPYWL 142
T+++T +IT T LF+ S+G + YL L ++I +GP+ L
Sbjct 869 THLVTKGLITYTYLFMSIFISIGCVCAYL-LSYYINGIGPWEL 910
> mmu:75698 Fam35a, 3110001K24Rik; family with sequence similarity
35, member A
Length=891
Score = 28.9 bits (63), Expect = 7.3, Method: Composition-based stats.
Identities = 16/68 (23%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 88 RRYLLLW--SSWGCTYVLTPAVITLTALFVYKESSLGSMALYLFLMFFITFVGPYWLIRS 145
++ ++LW +++G V +I LT + +Y++ +G L + +G Y ++
Sbjct 457 KKRVVLWRTAAFGALTVFLGDIILLTDVVLYEDQWIGETVLQSTFTSQLLNLGSYSYVQP 516
Query 146 QKYKNEIS 153
+KY N I+
Sbjct 517 EKYSNVIA 524
Lambda K H
0.327 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40