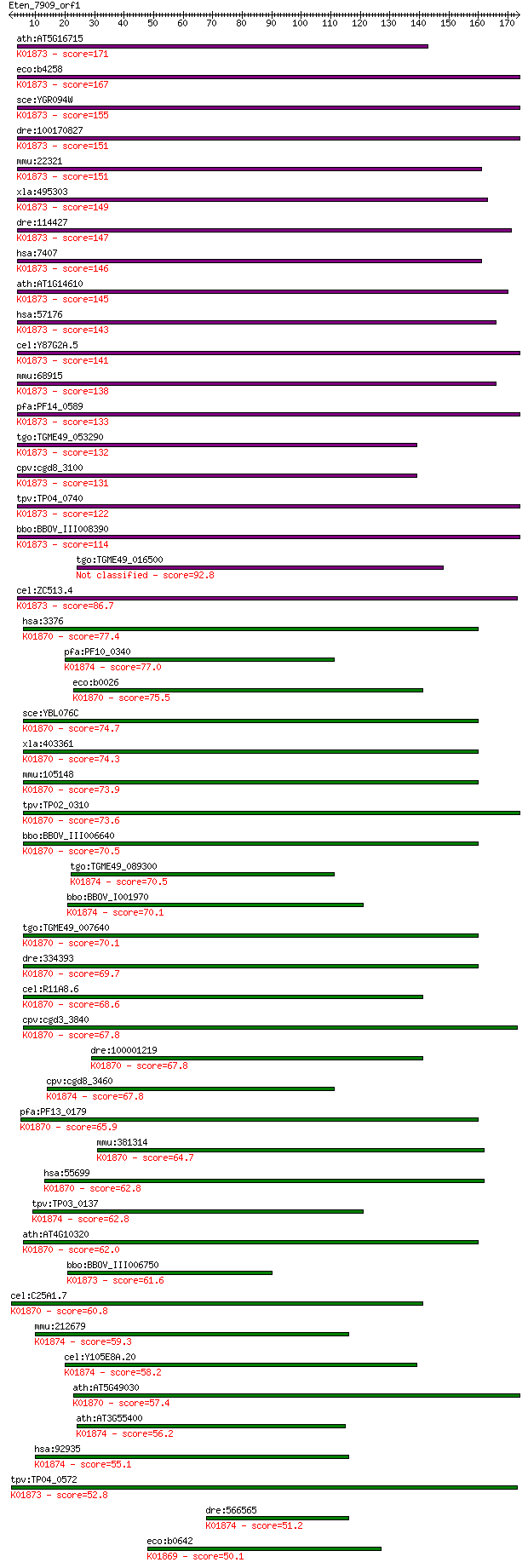
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7909_orf1
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding / a... 171 1e-42
eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1... 167 1e-41
sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA syn... 155 6e-38
dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase ... 151 1e-36
mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthe... 151 1e-36
xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9); K0... 149 4e-36
dre:114427 vars, CHUNP6879, im:7137378, vars1, vars2, wu:fb07a... 147 2e-35
hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.... 146 4e-35
ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRN... 145 7e-35
hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tR... 143 3e-34
cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2... 141 1e-33
mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA ... 138 1e-32
pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA ... 133 4e-31
tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9)... 132 4e-31
cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthe... 131 1e-30
tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl... 122 5e-28
bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.... 114 1e-25
tgo:TGME49_016500 tRNA synthetase, putative (EC:6.1.1.9 6.1.1.4) 92.8 5e-19
cel:ZC513.4 vrs-1; Valyl tRNA Synthetase family member (vrs-1)... 86.7 3e-17
hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; iso... 77.4 2e-14
pfa:PF10_0340 methionine-tRNA ligase, putative; K01874 methion... 77.0 3e-14
eco:b0026 ileS, ECK0027, ilvS, JW0024; isoleucyl-tRNA syntheta... 75.5 8e-14
sce:YBL076C ILS1; Ils1p (EC:6.1.1.5); K01870 isoleucyl-tRNA sy... 74.7 1e-13
xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870 i... 74.3 2e-13
mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04... 73.9 2e-13
tpv:TP02_0310 isoleucyl-tRNA synthetase; K01870 isoleucyl-tRNA... 73.6 3e-13
bbo:BBOV_III006640 17.m07588; isoleucyl-tRNA synthetase family... 70.5 2e-12
tgo:TGME49_089300 methionyl-tRNA synthetase, putative (EC:6.1.... 70.5 3e-12
bbo:BBOV_I001970 19.m02057; methionyl-tRNA synthetase (EC:6.1.... 70.1 4e-12
tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1... 70.1 4e-12
dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRN... 69.7 5e-12
cel:R11A8.6 irs-1; Isoleucyl tRNA Synthetase family member (ir... 68.6 9e-12
cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tR... 67.8 2e-11
dre:100001219 iars2, si:ch211-214p16.3, si:zc214p16.3; isoleuc... 67.8 2e-11
cpv:cgd8_3460 methionyl-tRNA synthetase of possible bacterial ... 67.8 2e-11
pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5); K... 65.9 7e-11
mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-... 64.7 1e-10
hsa:55699 IARS2, FLJ10326, ILERS; isoleucyl-tRNA synthetase 2,... 62.8 5e-10
tpv:TP03_0137 methionyl-tRNA synthetase (EC:6.1.1.10); K01874 ... 62.8 6e-10
ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine... 62.0 1e-09
bbo:BBOV_III006750 17.m07597; tRNA synthetases class I (I, L, ... 61.6 1e-09
cel:C25A1.7 irs-2; Isoleucyl tRNA Synthetase family member (ir... 60.8 2e-09
mmu:212679 Mars2, C730026E21Rik, MetRS; methionine-tRNA synthe... 59.3 6e-09
cel:Y105E8A.20 hypothetical protein; K01874 methionyl-tRNA syn... 58.2 1e-08
ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / ami... 57.4 3e-08
ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / ami... 56.2 5e-08
hsa:92935 MARS2, MetRS, mtMetRS; methionyl-tRNA synthetase 2, ... 55.1 1e-07
tpv:TP04_0572 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl... 52.8 6e-07
dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K018... 51.2 1e-06
eco:b0642 leuS, ECK0635, JW0637; leucyl-tRNA synthetase (EC:6.... 50.1 4e-06
> ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding /
aminoacyl-tRNA ligase/ nucleotide binding / valine-tRNA ligase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=970
Score = 171 bits (433), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 78/140 (55%), Positives = 102/140 (72%), Gaps = 2/140 (1%)
Query 4 LWPFETMGWPEQTA-ELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHG 62
LWPF T+GWP+ A + FYPT++L TG+DI+FFWVARMVMMG+EF +PF HV++HG
Sbjct 526 LWPFSTLGWPDVAAKDFNNFYPTNMLETGHDILFFWVARMVMMGIEFTGTVPFSHVYLHG 585
Query 63 LVRDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERVESARNFA 122
L+RDSQGRKMSKSLGN IDP++ I+ +G D LRF + G T G D+ ER+ + + F
Sbjct 586 LIRDSQGRKMSKSLGNVIDPLDTIKDFGTDALRFTIALG-TAGQDLNLSTERLTANKAFT 644
Query 123 NKLWNASRFMLMNLEGFDKT 142
NKLWNA +F+L +L T
Sbjct 645 NKLWNAGKFVLHSLPSLSDT 664
> eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=951
Score = 167 bits (424), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 90/213 (42%), Positives = 122/213 (57%), Gaps = 43/213 (20%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKD------IPFKH 57
LW F T+GWPE T L+QF+PTSV+V+G+DIIFFW+ARM+MM + F KD +PF
Sbjct 481 LWTFSTLGWPENTDALRQFHPTSVMVSGFDIIFFWIARMIMMTMHFIKDENGKPQVPFHT 540
Query 58 VFIHGLVRDSQGRKMSKSLGNGIDPVEV-------------------------------- 85
V++ GL+RD +G+KMSKS GN IDP+++
Sbjct 541 VYMTGLIRDDEGQKMSKSKGNVIDPLDMVDGISLPELLEKRTGNMMQPQLADKIRKRTEK 600
Query 86 -----IEKYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFD 140
IE +G D LRF L + G D+ + +R+E RNF NKLWNASRF+LMN EG D
Sbjct 601 QFPNGIEPHGTDALRFTLAALASTGRDINWDMKRLEGYRNFCNKLWNASRFVLMNTEGQD 660
Query 141 KTFVPEASDYTLADKWILSRYAKTAISITENLE 173
F +LAD+WIL+ + +T + E L+
Sbjct 661 CGFNGGEMTLSLADRWILAEFNQTIKAYREALD 693
> sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1104
Score = 155 bits (392), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 87/212 (41%), Positives = 119/212 (56%), Gaps = 45/212 (21%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
LWPF T+GWPE+T +++ FYP S+L TG+DI+FFWV RM+++GL+ +PFK VF H L
Sbjct 636 LWPFSTLGWPEKTKDMETFYPFSMLETGWDILFFWVTRMILLGLKLTGSVPFKEVFCHSL 695
Query 64 VRDSQGRKMSKSLGNGIDPVEVI------------------------------EKY---- 89
VRD+QGRKMSKSLGN IDP++VI E Y
Sbjct 696 VRDAQGRKMSKSLGNVIDPLDVITGIKLDDLHAKLLQGNLDPREVEKAKIGQKESYPNGI 755
Query 90 ---GADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVPE 146
G D +RF L T G D+ RVE R F NK++ A++F LM L G D + P
Sbjct 756 PQCGTDAMRFALCAYTTGGRDINLDILRVEGYRKFCNKIYQATKFALMRL-GDD--YQPP 812
Query 147 ASD-----YTLADKWILSRYAKTAISITENLE 173
A++ +L +KWIL + +T+ + E L+
Sbjct 813 ATEGLSGNESLVEKWILHKLTETSKIVNEALD 844
> dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1057
Score = 151 bits (381), Expect = 1e-36, Method: Composition-based stats.
Identities = 86/210 (40%), Positives = 104/210 (49%), Gaps = 42/210 (20%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+PF +GWP+QT +L QFYP S+L TG D+IFFWVARMVM+G E +PFK V H L
Sbjct 578 LFPFAMLGWPQQTKDLTQFYPNSILETGSDLIFFWVARMVMLGTELTGQLPFKQVLFHSL 637
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
VRD GRKMSKSLGN IDP++VI
Sbjct 638 VRDKYGRKMSKSLGNVIDPLDVISGVSLERLQEKVKEGNLDPRESIVAIEAQKKDFPKGI 697
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTF-VP 145
+ G D LRF L + G D+ V S R+F NK+W RF L L D + P
Sbjct 698 PECGTDALRFALCSYRAQGEDISLSMSHVLSCRHFCNKMWQTVRFTLATLN--DASLPTP 755
Query 146 EASDYTLA--DKWILSRYAKTAISITENLE 173
A Y L+ DKWI SR T + E
Sbjct 756 LAETYPLSSMDKWICSRLYSTVKECEQGFE 785
> mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1263
Score = 151 bits (381), Expect = 1e-36, Method: Composition-based stats.
Identities = 80/199 (40%), Positives = 107/199 (53%), Gaps = 45/199 (22%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+PF GWP Q+ +L FYP ++L TG+DI+FFWVARMVM+GL+ +PF+ V++H +
Sbjct 794 LFPFSIFGWPNQSEDLSVFYPGTLLETGHDILFFWVARMVMLGLKLTGKLPFREVYLHAI 853
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
VRD+ GRKMSKSLGN IDP++VI
Sbjct 854 VRDAHGRKMSKSLGNVIDPLDVIHGVSLQGLYDQLLNSNLDPSEVEKAKEGQKADFPAGI 913
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVPE 146
+ G D LRF L + G D+ R+ R+F NKLWNA++F L G K FVP
Sbjct 914 PECGTDALRFGLCAYTSQGRDINLDVNRILGYRHFCNKLWNATKFA---LRGLGKGFVPS 970
Query 147 ASDY-----TLADKWILSR 160
A+ +L D+WI SR
Sbjct 971 ATSKPEGHESLVDRWIRSR 989
> xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1243
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 80/198 (40%), Positives = 110/198 (55%), Gaps = 39/198 (19%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
++PF GWP QT +L+ FYP ++L TG+DI+FFWVARMVM+GL +PFK V++H +
Sbjct 774 IFPFSIFGWPNQTEDLQVFYPGTLLETGHDILFFWVARMVMLGLTLTGKLPFKEVYLHAV 833
Query 64 VRDSQGRKMSKSLGNGIDPVEVI----------------------------EKY------ 89
VRD+ GRKMSKSLGN IDP++VI +KY
Sbjct 834 VRDAHGRKMSKSLGNVIDPLDVINGITLEGLHKQLLESNLDPAELERAKDGQKYDYPNGI 893
Query 90 ---GADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNL-EGFDKT-FV 144
G D LRF L + G D+ R+ R+F NK+WNA++F + +L + F F
Sbjct 894 PECGTDALRFALCAYTSQGRDINLDVNRILGYRHFCNKIWNATKFAMRSLGDNFSPPDFA 953
Query 145 PEASDYTLADKWILSRYA 162
+LAD+WILSR +
Sbjct 954 GACGQESLADRWILSRLS 971
> dre:114427 vars, CHUNP6879, im:7137378, vars1, vars2, wu:fb07a11,
wu:fb07d10; valyl-tRNA synthetase; K01873 valyl-tRNA synthetase
[EC:6.1.1.9]
Length=1111
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 78/206 (37%), Positives = 113/206 (54%), Gaps = 40/206 (19%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
++PF GWP ++ +L+ FYP ++L TG+DI+FFWVARMVMMGL+ +PFK V++H +
Sbjct 638 IFPFSIFGWPNESEDLRVFYPGTLLETGHDILFFWVARMVMMGLKLTGKLPFKEVYLHAV 697
Query 64 VRDSQGRKMSKSLGNGIDPVEVI------------------------------------- 86
VRD+ GRKMSKSLGN IDP++VI
Sbjct 698 VRDAHGRKMSKSLGNVIDPLDVITGISLEGLYAQLADSNLDPLEIEKAKQGQKSDYPTGI 757
Query 87 EKYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNL-EGFDKTFVP 145
+ G D LRF L + G D+ R+ R+F NKLWNA +F + L EGF
Sbjct 758 PECGTDALRFALCAYTSQGRDINLDVNRILGYRHFCNKLWNAVKFAMRTLGEGFVPCEKA 817
Query 146 E-ASDYTLADKWILSRYAKTAISITE 170
+ +++D+WILSR + A+++ +
Sbjct 818 QLCGSESVSDRWILSRLS-AAVALCD 842
> hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1264
Score = 146 bits (368), Expect = 4e-35, Method: Composition-based stats.
Identities = 80/199 (40%), Positives = 107/199 (53%), Gaps = 45/199 (22%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+P +GWP Q+ +L FYP ++L TG+DI+FFWVARMVM+GL+ +PF+ V++H +
Sbjct 795 LFPLSILGWPNQSEDLSVFYPGTLLETGHDILFFWVARMVMLGLKLTGRLPFREVYLHAI 854
Query 64 VRDSQGRKMSKSLGNGIDPVEVI----------------------EKY------------ 89
VRD+ GRKMSKSLGN IDP++VI EK
Sbjct 855 VRDAHGRKMSKSLGNVIDPLDVIYGISLQGLHNQLLNSNLDPSEVEKAKEGQKADFPAGI 914
Query 90 ---GADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVPE 146
G D LRF L + G D+ R+ R+F NKLWNA++F L G K FVP
Sbjct 915 PECGTDALRFGLCAYMSQGRDINLDVNRILGYRHFCNKLWNATKFA---LRGLGKGFVPS 971
Query 147 ASDY-----TLADKWILSR 160
+ +L D+WI SR
Sbjct 972 PTSQPGGHESLVDRWIRSR 990
> ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / valine-tRNA ligase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1108
Score = 145 bits (366), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 84/206 (40%), Positives = 112/206 (54%), Gaps = 43/206 (20%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+P +GWP+ T + K FYPTSVL TG+DI+FFWVARMVMMG++ G ++PF V+ H +
Sbjct 628 LFPLSVLGWPDVTDDFKAFYPTSVLETGHDILFFWVARMVMMGMKLGGEVPFSKVYFHPM 687
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
+RD+ GRKMSKSLGN IDP+EVI
Sbjct 688 IRDAHGRKMSKSLGNVIDPLEVINGVTLEGLHKRLEEGNLDPKEVIVAKEGQVKDFPNGI 747
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNL-EGFD--KTF 143
+ G D LRF L++ + + RV R + NKLWNA RF +M L +G+ +T
Sbjct 748 PECGTDALRFALVSYTAQSDKINLDILRVVGYRQWCNKLWNAVRFAMMKLGDGYTPPQTL 807
Query 144 VPEASDYTLADKWILSRYAKTAISIT 169
PE ++ +WILS K AIS T
Sbjct 808 SPETMPFSC--QWILSVLNK-AISKT 830
> hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=923
Score = 143 bits (361), Expect = 3e-34, Method: Composition-based stats.
Identities = 77/203 (37%), Positives = 103/203 (50%), Gaps = 44/203 (21%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+PF +GWP++T +L +FYP S+L TG D++ FWV RMVM+G + +PF V +H +
Sbjct 451 LFPFSALGWPQETPDLARFYPLSLLETGSDLLLFWVGRMVMLGTQLTGQLPFSKVLLHPM 510
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
VRD QGRKMSKSLGN +DP ++I
Sbjct 511 VRDRQGRKMSKSLGNVLDPRDIISGVEMQVLQEKLRSGNLDPAELAIVAAAQKKDFPHGI 570
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVP- 145
+ G D LRF L + D+ V+S R+F NK+WNA RF+ L + FVP
Sbjct 571 PECGTDALRFTLCSHGVQAGDLHLSVSEVQSCRHFCNKIWNALRFI---LNALGEKFVPQ 627
Query 146 ---EASDYTLADKWILSRYAKTA 165
E S + D WILSR A A
Sbjct 628 PAEELSPSSPMDAWILSRLALAA 650
> cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1050
Score = 141 bits (355), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 80/212 (37%), Positives = 103/212 (48%), Gaps = 44/212 (20%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
+WPF GWP+ T ++ F+P +VL TG+DI+FFWVARMV M E +PFK + +H +
Sbjct 575 MWPFAVFGWPDATKDMDLFFPGAVLETGHDILFFWVARMVFMAQELTGKLPFKEILLHAM 634
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
+RD+ GRKMSKSLGN IDP++VI
Sbjct 635 IRDAHGRKMSKSLGNVIDPLDVIRGISLNDLQAQLLGGNLDEKEIAVAKEGQARDYPDGI 694
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDK----- 141
+ G D LRF L++ + G D+ RV R F NKLW RF L + DK
Sbjct 695 PECGVDALRFALLSYTSQGRDINLDVLRVHGYRKFCNKLWQVVRFALARIS--DKPEQKP 752
Query 142 TFVPEASDYTLADKWILSRYAKTAISITENLE 173
TF T D WILSR AK E L+
Sbjct 753 TFEINLKSATPTDLWILSRLAKAVKETNEALK 784
> mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=1060
Score = 138 bits (347), Expect = 1e-32, Method: Composition-based stats.
Identities = 77/203 (37%), Positives = 101/203 (49%), Gaps = 44/203 (21%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+PF +GWP +T +L FYP ++L TG D++ FWV RMVM+G + +PF V +H +
Sbjct 592 LFPFSALGWPRETPDLAHFYPLTLLETGSDLLMFWVGRMVMLGTQLTGQLPFSKVLLHSM 651
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
VRD QGRKMSKSLGN +DP ++I
Sbjct 652 VRDRQGRKMSKSLGNVLDPRDIISGQELQVLQAKLRDGNLDPGELAVAAAAQKKDFPYGI 711
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVP- 145
+ G D LRF L + G D+ V + R+F NKLWNA RF+L L FVP
Sbjct 712 PECGTDALRFALCSHGILGGDLHLSVSEVLNYRHFCNKLWNALRFVLRAL---GDNFVPQ 768
Query 146 ---EASDYTLADKWILSRYAKTA 165
E + + D WILSR A A
Sbjct 769 PAEEVTPSSPMDAWILSRLAFAA 791
> pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1090
Score = 133 bits (334), Expect = 4e-31, Method: Composition-based stats.
Identities = 75/213 (35%), Positives = 109/213 (51%), Gaps = 43/213 (20%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L PF ++GWP QT +L+ F+P ++L TG DI+FFWVARMVM+ L K +PFK +++H +
Sbjct 562 LVPFSSLGWPNQTEDLETFFPNTILETGQDILFFWVARMVMVSLHLMKKLPFKTIYLHAM 621
Query 64 VRDSQGRKMSKSLGNGIDPVEVIE------------------------------------ 87
+RDS+G KMSKS GN +DP+++I+
Sbjct 622 IRDSRGEKMSKSKGNVVDPLDIIDGISLNKLHEKLYEGNLPEKEIKRAIELQKKEFPKGI 681
Query 88 -KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNL-EGFDKTFV- 144
+ G D LRF L+T G ++ R+ R+F NKLWNA +F L L + +D +
Sbjct 682 PECGTDALRFGLLTYLKQGRNVNLDINRIIGYRHFCNKLWNAVKFFLKTLPDNYDNYNIL 741
Query 145 ---PE-ASDYTLADKWILSRYAKTAISITENLE 173
PE DKWIL + + +N E
Sbjct 742 IDKPEYVEKLQWEDKWILHKLNVYIKNANDNFE 774
> tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1042
Score = 132 bits (333), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 68/172 (39%), Positives = 91/172 (52%), Gaps = 37/172 (21%)
Query 4 LWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGL 63
L+PF GWP+ T +++ F+PTS+L TG+DI+FFWVARMVMM L+ +PF VF+H +
Sbjct 510 LFPFSVFGWPDNTEDIQAFFPTSLLETGHDILFFWVARMVMMSLQLTDKLPFDTVFLHAM 569
Query 64 VRDSQGRKMSKSLGNGIDPVEV-------------------------------------I 86
VRD+ G+KMSKS GN IDP+EV I
Sbjct 570 VRDAHGQKMSKSKGNVIDPLEVISGISLQDLQAKLHKGNLPEKEIKRAEEVLKKEFPKGI 629
Query 87 EKYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEG 138
E G D LR L+ G ++ RV R+F NKLWNA++F + E
Sbjct 630 EACGCDALRLGLLAYTRQGRNVNLDLNRVVGYRHFCNKLWNATKFAMDKFEA 681
> cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthetase
[EC:6.1.1.9]
Length=1042
Score = 131 bits (330), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 69/178 (38%), Positives = 94/178 (52%), Gaps = 43/178 (24%)
Query 4 LWPFETMGWPEQTAE------LKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKH 57
L PF +GWP++ + L F+PT++L TG DI+FFWVARMVM+ +PFK
Sbjct 541 LIPFSPLGWPDEVNQKTKLNDLSTFFPTTLLETGSDILFFWVARMVMLSFACTDKLPFKT 600
Query 58 VFIHGLVRDSQGRKMSKSLGNGIDPVEVIE------------------------------ 87
+++H +VRDSQGRKMSKSLGN IDP+E+IE
Sbjct 601 IYLHAMVRDSQGRKMSKSLGNVIDPIEIIEGISLDDLNKKLDQGNLPLQEIKKSKENNLK 660
Query 88 -------KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEG 138
+ GAD LR L+ G ++ +RV + R+F NKLWNA +F N+E
Sbjct 661 DFPDGIPECGADALRIGLLAYTKQGRNINLDTKRVVAYRHFCNKLWNACKFAFGNIEN 718
> tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1010
Score = 122 bits (307), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 73/224 (32%), Positives = 108/224 (48%), Gaps = 54/224 (24%)
Query 4 LWPFETMGWPE-QTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHG 62
L+P T+GWP+ T++ K F+PTS+L TG DIIFFWVARMVM+ L F +PF +++H
Sbjct 516 LFPLSTLGWPDTDTSDFKSFFPTSLLETGNDIIFFWVARMVMLSLHFVDMLPFNEIYMHP 575
Query 63 LVRDSQGRKMSKSLGNGIDPVEVIEK---------------------------------- 88
LVRDS+G KMSKS GN +DP+++IE
Sbjct 576 LVRDSRGEKMSKSKGNVVDPIDIIEGTTLERLNQNILNSSLPQGEIKRALALQKQQFPDG 635
Query 89 ---YGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDKTFVP 145
G D LR L+ + ++ S+R+F NK+WNA++F ++ + F +
Sbjct 636 IPICGVDGLRLGLLALMRHNRAILLDVNKLVSSRHFGNKIWNATKFAILRTKFFRPSVQH 695
Query 146 EASDYTLA----------------DKWILSRYAKTAISITENLE 173
+ Y DKWIL + + +T+ LE
Sbjct 696 TYNHYNTVKSYKGDNSLECKFKWEDKWILHKLNQYTKRVTDGLE 739
> bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=972
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 70/210 (33%), Positives = 102/210 (48%), Gaps = 43/210 (20%)
Query 4 LWPFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHG 62
L+P T+GWP E+ + +F+PT++L TG DIIFFWVARMVMM L +PF V++H
Sbjct 500 LFPLSTLGWPDEKHPDFMKFFPTTLLETGNDIIFFWVARMVMMSLHLVGKLPFNTVYLHP 559
Query 63 LVRDSQGRKMSKSLGNGIDPVEVIE----------------------------------- 87
LVRD++G KMSKS GN +DP+EVIE
Sbjct 560 LVRDARGEKMSKSKGNVLDPLEVIEGATLDSLIDKINNSSLPQGEIKKAIVLQKQQFPQG 619
Query 88 --KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEG--FDKTF 143
G D LR L++ + ++ +++F NK+WNA +F + + D
Sbjct 620 IPACGTDALRLGLLSLTRQNRAILLDVNKIICSKHFGNKIWNALKFTMPKITQVPMDINS 679
Query 144 VPEASDYTLADKWILSRYAKTAISITENLE 173
VP D+W+LS+ + +T E
Sbjct 680 VPCLH---WEDRWVLSKLNEYISKVTAAFE 706
> tgo:TGME49_016500 tRNA synthetase, putative (EC:6.1.1.9 6.1.1.4)
Length=1762
Score = 92.8 bits (229), Expect = 5e-19, Method: Composition-based stats.
Identities = 51/130 (39%), Positives = 75/130 (57%), Gaps = 14/130 (10%)
Query 24 PTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDPV 83
P + LVTG DI+FFWV R ++ + PF V +HGL+ D +G+K+SK+ GN V
Sbjct 1237 PQTELVTGEDILFFWVVRQFVLCAALTQQAPFSRVTLHGLLVDREGKKLSKTKGN----V 1292
Query 84 E------VIEKYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLE 137
E +I++ GAD LR+ ++G +PG + F + S+R F +KLW+A RF +E
Sbjct 1293 ERGYLDRLIDEAGADALRWTFLSGMSPGASVAFDEGALASSRRFLHKLWHAGRF----IE 1348
Query 138 GFDKTFVPEA 147
F F P A
Sbjct 1349 RFASQFPPPA 1358
> cel:ZC513.4 vrs-1; Valyl tRNA Synthetase family member (vrs-1);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=918
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 62/212 (29%), Positives = 94/212 (44%), Gaps = 51/212 (24%)
Query 4 LWPFETMGW----PEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGK-DIPFKHV 58
L P GW P QT L +V+ TG+DI FWVARM+ + ++ + PF V
Sbjct 446 LVPLVKNGWLEGAPIQTPSL------NVMETGWDISGFWVARMIALNMKLSSGETPFGKV 499
Query 59 FIHGLVRDSQGRKMSKSLGNGIDPVEV--------------------------------- 85
+HGLVRD++GRKMSKSLGN IDP++V
Sbjct 500 VLHGLVRDNEGRKMSKSLGNVIDPLDVLDGITFEKMIERVKSSAHEKEEIDNAVKDLTKR 559
Query 86 ----IEKYGADTLRFMLITGNTPGNDMRFYWERVE-SARNFANKLWNASRFMLMNLEGFD 140
I + G D LRF L+ + D+ +F NK+WN + + E +
Sbjct 560 FPNGISRCGPDALRFALLKYDVLSTDIPIDVSNAAIEGLHFCNKIWNLAAYYDQLAEKCE 619
Query 141 KTFVPEASDYTLADKWILSRYAKTAISITENL 172
+ + L D+WI++R + T ++ ++
Sbjct 620 --VIKDVDSDRLVDEWIMARLSTTVSTVDAHM 649
> hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1262
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 55/169 (32%), Positives = 83/169 (49%), Gaps = 19/169 (11%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P E E + +P + G D W ++++ PFK+V ++GLV
Sbjct 534 PYAQVHYPFENKREFEDAFPADFIAEGIDQTRGWFYTLLVLATALFGQPPFKNVIVNGLV 593
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESA-RN 120
S G+KMSK N DPV +I+KYGAD LR LI N+P ++RF E V ++
Sbjct 594 LASDGQKMSKRKKNYPDPVSIIQKYGADALRLYLI--NSPVVRAENLRFKEEGVRDVLKD 651
Query 121 FANKLWNASRFMLMNLEGFDK----------TFVPEASDYTLADKWILS 159
+NA RF++ N+ K V E+ + T D+WILS
Sbjct 652 VLLPWYNAYRFLIQNVLRLQKEEEIEFLYNENTVRESPNIT--DRWILS 698
> pfa:PF10_0340 methionine-tRNA ligase, putative; K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=889
Score = 77.0 bits (188), Expect = 3e-14, Method: Composition-based stats.
Identities = 39/92 (42%), Positives = 56/92 (60%), Gaps = 3/92 (3%)
Query 20 KQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPF-KHVFIHGLVRDSQGRKMSKSLGN 78
K+++P +V + G DI W ++ + +IP K VF HG V + G+KMSKSLGN
Sbjct 465 KKYWPANVHIIGKDIT--WFHTVIFPTILLSANIPLSKSVFSHGFVLAADGKKMSKSLGN 522
Query 79 GIDPVEVIEKYGADTLRFMLITGNTPGNDMRF 110
I+P+E+I+KYGAD R+ +I G D RF
Sbjct 523 VINPLEIIDKYGADAFRYHVIKETKRGCDTRF 554
> eco:b0026 ileS, ECK0027, ilvS, JW0024; isoleucyl-tRNA synthetase
(EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=938
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/118 (33%), Positives = 62/118 (52%), Gaps = 1/118 (0%)
Query 23 YPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDP 82
+ + + G D W +M+ P++ V HG D QGRKMSKS+GN + P
Sbjct 554 HAADMYLEGSDQHRGWFMSSLMISTAMKGKAPYRQVLTHGFTVDGQGRKMSKSIGNTVSP 613
Query 83 VEVIEKYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFD 140
+V+ K GAD LR + + + G +M E ++ A + ++ N +RF+L NL GFD
Sbjct 614 QDVMNKLGADILRLWVASTDYTG-EMAVSDEILKRAADSYRRIRNTARFLLANLNGFD 670
> sce:YBL076C ILS1; Ils1p (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1072
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 81/168 (48%), Gaps = 19/168 (11%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P E T + + P + + G D W + ++G +P+K+V + G+V
Sbjct 536 PYASQHYPFENTEKFDERVPANFISEGLDQTRGWFYTLAVLGTHLFGSVPYKNVIVSGIV 595
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESARNF 121
+ GRKMSKSL N DP V+ KYGAD LR LI N+P ++F + E +
Sbjct 596 LAADGRKMSKSLKNYPDPSIVLNKYGADALRLYLI--NSPVLKAESLKF---KEEGVKEV 650
Query 122 ANKL----WNASRF------MLMNLEGFDKTFVPEASDYTLADKWILS 159
+K+ WN+ +F +L + D + + D+WIL+
Sbjct 651 VSKVLLPWWNSFKFLDGQIALLKKMSNIDFQYDDSVKSDNVMDRWILA 698
> xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1259
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/169 (31%), Positives = 84/169 (49%), Gaps = 19/169 (11%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P E E + +P + G D W ++++ PFK+V ++GLV
Sbjct 534 PYAQVHYPFENRREFEDCFPADFIAEGIDQTRGWFYTLLVLSSALFGQPPFKNVIVNGLV 593
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESARNF 121
S G+KMSKS N DP++VI YG+D LR LI N+P ++RF E V
Sbjct 594 LASDGQKMSKSKKNYPDPMQVINVYGSDALRLYLI--NSPVVRAENLRFKEEGVRDVLKD 651
Query 122 ANKLW-NASRFMLMNLEG----------FDKTFVPEASDYTLADKWILS 159
W NA RF++ N+ F+++ ++S+ + DKWI+S
Sbjct 652 VFLPWYNAYRFLMQNIYRLHKEEDIHFLFNESIKQQSSN--IMDKWIIS 698
> mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04Rik,
ILRS; isoleucine-tRNA synthetase (EC:6.1.1.5); K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1262
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 52/169 (30%), Positives = 84/169 (49%), Gaps = 19/169 (11%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P E E + +P + G D W ++++ PFK+V ++GL+
Sbjct 534 PYAQVHYPFESKREFEDAFPADFIAEGIDQTRGWFYTLLVLATALFGQPPFKNVIVNGLI 593
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESA-RN 120
S G+KMSK N DPV +I+KYGAD LR LI N+P ++RF E V ++
Sbjct 594 LASDGQKMSKRKKNYPDPVSIIDKYGADALRLYLI--NSPVVRAENLRFKEEGVRDVLKD 651
Query 121 FANKLWNASRFMLMNLEG----------FDKTFVPEASDYTLADKWILS 159
+NA RF + N+ +++ V E+ + T D+W+LS
Sbjct 652 VLLPWYNAYRFFIQNVFRLHKEEEVKFLYNEHTVRESPNIT--DRWVLS 698
> tpv:TP02_0310 isoleucyl-tRNA synthetase; K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1129
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 54/183 (29%), Positives = 90/183 (49%), Gaps = 19/183 (10%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWV-ARMVMMGLEFGKDIPFKHVFIHGL 63
P + +P E L Q +P + + G D W MV+ L F K PFK+V ++GL
Sbjct 604 PLAKIHYPFENKDSLDQHFPANFIAEGLDQTRGWFYTLMVLSTLLFNKS-PFKNVIVNGL 662
Query 64 VRDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITG-NTPGNDMRFYWERVESARNFA 122
+ S G+KMSK L N DP+++I ++GAD+LR LI+ +RF +S RN
Sbjct 663 ILASDGKKMSKRLKNYPDPLDIINQFGADSLRLFLISSPAVKAEPVRF---STDSVRNVL 719
Query 123 NKL----WNASRFMLMNLEGFDKT----FVPEA----SDYTLADKWILSRYAKTAISITE 170
+ +++ RF++ + ++ T FVP+ + D+WI + + S+
Sbjct 720 KDVILPWFHSYRFLVQEVTRYEVTHKMKFVPDPEAPFKSSCVMDRWINTITQELIYSVHH 779
Query 171 NLE 173
+E
Sbjct 780 EME 782
> bbo:BBOV_III006640 17.m07588; isoleucyl-tRNA synthetase family
protein (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1080
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/165 (29%), Positives = 78/165 (47%), Gaps = 11/165 (6%)
Query 6 PFETMGWPEQTAEL-KQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ M +P + EL + +P + G D W ++++ F+++ ++GLV
Sbjct 553 PYAKMHYPFENKELFTEIFPADFVAEGLDQTRGWFYTLMVISTHLFNAPAFRNIIVNGLV 612
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGN-TPGNDMRFYWERVESARNFAN 123
S G+KMSK L N DPVEVI +YGAD++R LI+ +RF E V
Sbjct 613 LASDGKKMSKRLRNFADPVEVINQYGADSVRLYLISSPVVRAEPLRFVTEGVRGILKDVI 672
Query 124 KLW-NASRFMLMNLEGFD----KTFVPEA----SDYTLADKWILS 159
W +A RF++ ++ F+P + Y + D W+ S
Sbjct 673 LPWFHAYRFLVQEASRYEMVTGNRFIPSTEITMNSYCVMDLWLHS 717
> tgo:TGME49_089300 methionyl-tRNA synthetase, putative (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=976
Score = 70.5 bits (171), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 53/90 (58%), Gaps = 3/90 (3%)
Query 22 FYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPF-KHVFIHGLVRDSQGRKMSKSLGNGI 80
F+P V + G DI+ W ++ + D+P K VF HG V + G KMSKSLGN +
Sbjct 521 FWPADVHIIGKDIV--WFHTVIWPCMLMAADLPLPKCVFGHGFVTAADGEKMSKSLGNVV 578
Query 81 DPVEVIEKYGADTLRFMLITGNTPGNDMRF 110
DP ++++ + AD+ RF L+ G+D+RF
Sbjct 579 DPNDILQAHDADSFRFYLVRETKFGSDLRF 608
> bbo:BBOV_I001970 19.m02057; methionyl-tRNA synthetase (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=534
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 61/103 (59%), Gaps = 7/103 (6%)
Query 21 QFYPTSVLVTGYDIIFFWVA--RMVMMGLEFGKDIPF-KHVFIHGLVRDSQGRKMSKSLG 77
+ +P + V G DII+F+ ++M LE +P K +F HG + + GRKMSKSLG
Sbjct 274 KLWPPDLQVIGKDIIWFFAVIWTSILMSLE----VPLPKTIFAHGFITAADGRKMSKSLG 329
Query 78 NGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERVESARN 120
N ++ E+++KYG DTLR+ +I + G+D++F + N
Sbjct 330 NVVNTKELLDKYGVDTLRYYMIRDSVFGSDVKFDLSSLADLHN 372
> tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1103
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 47/171 (27%), Positives = 86/171 (50%), Gaps = 22/171 (12%)
Query 6 PFETMGWPEQTAE-LKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P + AE ++ +P + G D W ++++ PF+++ ++GLV
Sbjct 561 PYGQIHYPFENAETFEKGFPADFIAEGLDQTRGWFYTLMVISTALFDRPPFQNLIVNGLV 620
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESARNF 121
+ GRKMSK L N DP EV+ +GAD LR L+ N+P +RF R+E ++
Sbjct 621 LAADGRKMSKRLKNYPDPTEVVHAHGADALRVYLV--NSPVVKAEALRF---RIEGVKDV 675
Query 122 ANKL----WNASRFMLMNLEGF----DKTFVPEASDYTLA-----DKWILS 159
+ ++A RF++ + + D+ F P + ++ D+WI+S
Sbjct 676 VKDVFLPWFHACRFLIQEVIRYESAGDRKFEPCKASALISKGNSMDRWIIS 726
> dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase
[EC:6.1.1.5]
Length=1271
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 52/167 (31%), Positives = 79/167 (47%), Gaps = 15/167 (8%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P E E + +P + G D W ++++ PFK+V ++GLV
Sbjct 534 PYAQVHYPFENRREFEDAFPADFIAEGIDQTRGWFYTLLVLSTALFGKPPFKNVIVNGLV 593
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESARNF 121
S G+KMSK N DP +++ YGAD LR LI N+P ++RF E V
Sbjct 594 LASDGQKMSKRKKNYPDPGLIVQSYGADALRLYLI--NSPVVRAENLRFKEEGVRDVLKD 651
Query 122 ANKLW-NASRFMLMNL------EGFDKTFVPEASDYT--LADKWILS 159
W NA RF++ N+ EG + + S + + DKWI S
Sbjct 652 VFLPWYNAYRFLVQNVQRLQKEEGVEFLYNERTSSVSNNIMDKWIQS 698
> cel:R11A8.6 irs-1; Isoleucyl tRNA Synthetase family member (irs-1);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1141
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 45/143 (31%), Positives = 71/143 (49%), Gaps = 13/143 (9%)
Query 6 PFETMGWP-EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ +P E + +P + G D W ++++ PFK++ +GLV
Sbjct 535 PYAQNHYPFENRKIFEDNFPADFIAEGIDQTRGWFYTLLVLSTALFNKPPFKNLICNGLV 594
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESARNF 121
S G KMSKS N DP+ ++ KYGAD LR LI N+P G ++RF R E R+
Sbjct 595 LASDGAKMSKSKKNYPDPMLIVNKYGADALRLYLI--NSPVVRGENLRF---REEGVRDL 649
Query 122 ANKL----WNASRFMLMNLEGFD 140
+ +NA RF + N++ ++
Sbjct 650 LKDVFLPWFNAYRFFVQNVQAYE 672
> cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1100
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 50/172 (29%), Positives = 81/172 (47%), Gaps = 18/172 (10%)
Query 6 PFETMGWPEQTAEL-KQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ ++ +P + EL +P + G D W ++++ FK++ ++GLV
Sbjct 547 PYASINYPFENQELFNNIFPADFVGEGLDQTRGWFYTLMVLSTALFDQPAFKNIIVNGLV 606
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVESA-RN 120
S G+KMSK L N DP++++ KYGAD+LR LI N+P +RF E V+ ++
Sbjct 607 LASDGKKMSKRLKNYPDPLDIVNKYGADSLRLFLI--NSPVVRAETLRFKEEGVKDVIKD 664
Query 121 FANKLWNASRFMLMNLEGFDKTFVPEASDYTLADKWILSRYAKTAISITENL 172
++ RF FV EA Y + I + K I T N+
Sbjct 665 VLLPWYHCYRF-----------FVQEAIRYENTNHNIKFKSNKQVIKNTNNI 705
> dre:100001219 iars2, si:ch211-214p16.3, si:zc214p16.3; isoleucine-tRNA
synthetase 2, mitochondrial (EC:6.1.1.5); K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=983
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 62/121 (51%), Gaps = 10/121 (8%)
Query 29 VTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDPVEVIE- 87
V G D I W ++ + P+K + +HG +G KMSKS+GN +DP VI
Sbjct 591 VEGKDQIGGWFQSSLLTSVAVRNKAPYKALVVHGFAVSEKGEKMSKSIGNVVDPDVVING 650
Query 88 --------KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGF 139
YGAD LR+ + N ++++ + +A++ NKL N +F+L NL+GF
Sbjct 651 GKDSSVSPPYGADVLRWWVAESNV-FSEVQIGPNALNAAKDSINKLRNTLKFLLGNLQGF 709
Query 140 D 140
D
Sbjct 710 D 710
> cpv:cgd8_3460 methionyl-tRNA synthetase of possible bacterial
origin ; K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=581
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 54/97 (55%), Gaps = 1/97 (1%)
Query 14 EQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMS 73
E+ + +++ +V + G DI +F M L G +P K +F HG V G+KMS
Sbjct 281 EENSLFNKYWENTVHIVGKDITWFHSVIWPAMLLSVGLSLP-KTIFAHGFVTAPDGKKMS 339
Query 74 KSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRF 110
KSLGN +DP E I+K G+D R+ L GND+++
Sbjct 340 KSLGNVVDPFEQIDKIGSDPFRYYLAREGRFGNDIKY 376
> pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1272
Score = 65.9 bits (159), Expect = 7e-11, Method: Composition-based stats.
Identities = 46/165 (27%), Positives = 80/165 (48%), Gaps = 15/165 (9%)
Query 5 WPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
+PF T +T + + +P + G D W ++++ PFK++ +GLV
Sbjct 722 YPFST-----ETNDFHKIFPADFIAEGLDQTRGWFYTLLVISTLLFNKAPFKNLICNGLV 776
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITG-NTPGNDMRFYWERV-ESARNFA 122
S G+KMSK L N DP+ ++ KYGAD+LR LI +++F + V E ++F
Sbjct 777 LASDGKKMSKRLKNYPDPLYILNKYGADSLRLYLINSVAVRAENLKFQEKGVNEIVKSFI 836
Query 123 NKLWNASRFMLMNLEGFDKTFVPE---ASDY-----TLADKWILS 159
+++ RF + ++ T + +DY + D+WI S
Sbjct 837 LPYYHSFRFFSQEVTRYETTNKMQFLFNTDYIYKNDNIMDQWIFS 881
> mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-tRNA
synthetase 2, mitochondrial (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1012
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 67/140 (47%), Gaps = 14/140 (10%)
Query 31 GYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDPVEVIE--- 87
G D + W ++ + PF+ V +HG +G KMSKSLGN I+P +I
Sbjct 624 GKDQLGGWFQSSLLTSVATRSKAPFRTVMVHGFTLGEKGEKMSKSLGNVINPDTIISGGK 683
Query 88 ------KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFMLMNLEGFDK 141
YGAD LR+ + N ++ + +AR+ +KL N RF+L NL GF+
Sbjct 684 DHSKEPPYGADILRWWIAESNV-FTEVTIGPSVLSAARDDISKLRNTLRFLLGNLTGFN- 741
Query 142 TFVPEASDYTLADKWILSRY 161
PE + + +++ +Y
Sbjct 742 ---PETDSVPVKNMYVIDQY 758
> hsa:55699 IARS2, FLJ10326, ILERS; isoleucyl-tRNA synthetase
2, mitochondrial (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase
[EC:6.1.1.5]
Length=1012
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 73/158 (46%), Gaps = 22/158 (13%)
Query 13 PEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKM 72
P+Q A+L + G D + W ++ + K P+K V +HG +G KM
Sbjct 614 PDQRADL--------YLEGKDQLGGWFQSSLLTSVAARKRAPYKTVIVHGFTLGEKGEKM 665
Query 73 SKSLGNGIDPVEVIE---------KYGADTLRFMLITGNTPGNDMRFYWERVESARNFAN 123
SKSLGN I P V+ YGAD LR+ + N ++ + +AR+ +
Sbjct 666 SKSLGNVIHPDVVVNGGQDQSKEPPYGADVLRWWVADSNV-FTEVAIGPSVLNAARDDIS 724
Query 124 KLWNASRFMLMNLEGFDKTFVPEASDYTLADKWILSRY 161
KL N RF+L N+ F+ PE + D +++ +Y
Sbjct 725 KLRNTLRFLLGNVADFN----PETDSIPVNDMYVIDQY 758
> tpv:TP03_0137 methionyl-tRNA synthetase (EC:6.1.1.10); K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=691
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 63/113 (55%), Gaps = 4/113 (3%)
Query 9 TMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPF-KHVFIHGLVRDS 67
+ G E +E+ ++PT + V G DI W +++ + F +P + + HG ++ S
Sbjct 420 SSGLGESLSEM-TYWPTDLQVIGKDIS--WFQSVILATMCFSMGLPLTRKILAHGFIQGS 476
Query 68 QGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERVESARN 120
GRKMSKSLGN ++ E++++ G ++ R+ L+ G+D+ F +++ N
Sbjct 477 DGRKMSKSLGNVLNLDELLQEVGVESFRYYLLKSTVMGSDVNFDQNQLKEMHN 529
> ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine--tRNA
ligase, putative (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1190
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 48/167 (28%), Positives = 81/167 (48%), Gaps = 15/167 (8%)
Query 6 PFETMGWPEQTAEL-KQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLV 64
P+ + +P + EL ++ +P + G D W ++++ + FK++ +GLV
Sbjct 566 PYAYIHYPFENKELFEKNFPGDFVAEGLDQTRGWFYTLMVLSTALFEKPAFKNLICNGLV 625
Query 65 RDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTP---GNDMRFYWERVES-ARN 120
G+KM+K L N P+EVI++YGAD +R LI N+P +RF E V ++
Sbjct 626 LAEDGKKMAKKLRNYPPPLEVIDEYGADAVRLYLI--NSPVVRAEPLRFKKEGVLGVVKD 683
Query 121 FANKLWNASRFMLMNLEGFDKT----FVP----EASDYTLADKWILS 159
+NA RF++ N + + FVP + D+WI S
Sbjct 684 VFLPWYNAYRFLVQNAKRLETEGGVPFVPTDLATIQSANILDQWIHS 730
> bbo:BBOV_III006750 17.m07597; tRNA synthetases class I (I, L,
M and V) family protein; K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=809
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 45/70 (64%), Gaps = 1/70 (1%)
Query 21 QFYPTSVLVTGYDIIFFWVARMVMMGLEFGKD-IPFKHVFIHGLVRDSQGRKMSKSLGNG 79
Q T +L TGYDI+ WVARM+ + + + +PF + +HG++ D++G+KMSKS GN
Sbjct 476 QLDVTRLLYTGYDILHSWVARMLALCTKLNESRLPFDKLVLHGIINDTKGKKMSKSWGNT 535
Query 80 IDPVEVIEKY 89
+ + I+ +
Sbjct 536 VTATQFIQSF 545
> cel:C25A1.7 irs-2; Isoleucyl tRNA Synthetase family member (irs-2);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=970
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/147 (23%), Positives = 68/147 (46%), Gaps = 8/147 (5%)
Query 2 VDLWPFETMGW-PEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFI 60
+D+W + W + + ++ + V++ G D W +++ + +P+K + +
Sbjct 588 MDVWLDSGLAWHAARDNDTEREHVADVVLEGVDQFRGWFQSLLLTSVAVQNKLPYKKIIV 647
Query 61 HGLVRDSQGRKMSKSLGNGIDPVEVIE------KYGADTLRF-MLITGNTPGNDMRFYWE 113
HG D KMSKS+GN +DP + + GAD LRF + ++G+ + + +
Sbjct 648 HGFCIDENNNKMSKSIGNVVDPTMLTDGSLKQKAIGADGLRFWVALSGSENAGESKIGMK 707
Query 114 RVESARNFANKLWNASRFMLMNLEGFD 140
++ N RFM+ +GFD
Sbjct 708 IIDDVDKKVIAFRNGFRFMIGGCQGFD 734
> mmu:212679 Mars2, C730026E21Rik, MetRS; methionine-tRNA synthetase
2 (mitochondrial) (EC:6.1.1.10); K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=586
Score = 59.3 bits (142), Expect = 6e-09, Method: Composition-based stats.
Identities = 36/110 (32%), Positives = 59/110 (53%), Gaps = 12/110 (10%)
Query 10 MGWPEQTAELKQFYPTSVLVTGYDIIFF----WVARMVMMGLEFGKDIPFKHVFIHGLVR 65
+G+P+ A+ K ++P + + G DI+ F W A ++ GL P +++H
Sbjct 283 VGYPD--ADFKSWWPATSHIIGKDILKFHAIYWPALLLGAGLR-----PPHRIYVHSHWT 335
Query 66 DSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERV 115
S G+KMSKSLGN +DP +++Y D R+ L+ P D +Y E+V
Sbjct 336 VS-GQKMSKSLGNVVDPRTCLDRYTVDGFRYFLLRQGVPNWDCDYYDEKV 384
> cel:Y105E8A.20 hypothetical protein; K01874 methionyl-tRNA synthetase
[EC:6.1.1.10]
Length=524
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 44/130 (33%), Positives = 62/130 (47%), Gaps = 19/130 (14%)
Query 20 KQFYPTSVLVTGYDI----IFFWVARMVMMGLEFGKDIPFKHVFIHG--LVRDSQGRKMS 73
+ +P + V G DI +++W A ++ GL +P K VFIHG L+ + KMS
Sbjct 262 QSVWPPTCQVIGKDITKFHLYYWPAFLMAAGL----SLPQK-VFIHGHWLIDNV---KMS 313
Query 74 KSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERV-----ESARNFANKLWNA 128
KSLGN ++P E I K+ A+ LR+ L+ P D F W N L N
Sbjct 314 KSLGNVVNPSEAITKFTAEGLRYFLLKHGNPSYDCSFNWNSCLQTINSDLVNNVGNLLNR 373
Query 129 SRFMLMNLEG 138
S +N EG
Sbjct 374 STVEKINKEG 383
> ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / aminoacyl-tRNA
ligase/ catalytic/ isoleucine-tRNA ligase/ nucleotide
binding; K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=955
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 44/160 (27%), Positives = 67/160 (41%), Gaps = 10/160 (6%)
Query 23 YPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDP 82
+P V + G D W ++ + P+ V HG V D +G KMSKSLGN +DP
Sbjct 675 FPADVYLEGTDQHRGWFQSSLLTSIATQGKAPYSAVITHGFVLDEKGMKMSKSLGNVVDP 734
Query 83 VEVIE---------KYGADTLRFMLITGNTPGNDMRFYWERVESARNFANKLWNASRFML 133
VIE YGAD +R + + + G D+ + + + KL R++L
Sbjct 735 RLVIEGGKNSKDAPAYGADVMRLWVSSVDYTG-DVLIGPQILRQMSDIYRKLRGTLRYLL 793
Query 134 MNLEGFDKTFVPEASDYTLADKWILSRYAKTAISITENLE 173
NL + D + D+ L + +I E E
Sbjct 794 GNLHDWRVDNAVPYQDLPIIDQHALFQLENVVKNIQECYE 833
> ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / aminoacyl-tRNA
ligase/ methionine-tRNA ligase/ nucleotide binding;
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=533
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 52/92 (56%), Gaps = 4/92 (4%)
Query 24 PTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIHG-LVRDSQGRKMSKSLGNGIDP 82
P S+ + G DI+ F M + G ++P K VF HG L +D G KM KSLGN ++P
Sbjct 321 PASLHLIGKDILRFHAVYWPAMLMSAGLELP-KMVFGHGFLTKD--GMKMGKSLGNTLEP 377
Query 83 VEVIEKYGADTLRFMLITGNTPGNDMRFYWER 114
E+++K+G D +R+ + GND + +R
Sbjct 378 FELVQKFGPDAVRYFFLREVEFGNDGDYSEDR 409
> hsa:92935 MARS2, MetRS, mtMetRS; methionyl-tRNA synthetase 2,
mitochondrial (EC:6.1.1.10); K01874 methionyl-tRNA synthetase
[EC:6.1.1.10]
Length=593
Score = 55.1 bits (131), Expect = 1e-07, Method: Composition-based stats.
Identities = 35/110 (31%), Positives = 56/110 (50%), Gaps = 12/110 (10%)
Query 10 MGWPEQTAELKQFYPTSVLVTGYDIIFF----WVARMVMMGLEFGKDIPFKHVFIHGLVR 65
+G+P AE K ++P + + G DI+ F W A ++ G+ P + + +H
Sbjct 290 IGYP--NAEFKSWWPATSHIIGKDILKFHAIYWPAFLLGAGMS-----PPQRICVHSHWT 342
Query 66 DSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERV 115
G+KMSKSLGN +DP + +Y D R+ L+ P D +Y E+V
Sbjct 343 VC-GQKMSKSLGNVVDPRTCLNRYTVDGFRYFLLRQGVPNWDCDYYDEKV 391
> tpv:TP04_0572 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=820
Score = 52.8 bits (125), Expect = 6e-07, Method: Composition-based stats.
Identities = 50/198 (25%), Positives = 85/198 (42%), Gaps = 30/198 (15%)
Query 2 VDLWPFETMGWPEQTAELKQFYPTSVLVTGYDIIFFWVARMVMMGLEFGKDIPFKHVFIH 61
+D W F + WP T S+L T YDI+ W+ +M+++ ++ F V++H
Sbjct 485 LDTW-FTSSLWPMIT-HTNTPVTRSILFTCYDILENWIVKMLLICSNIDENKLFSEVYLH 542
Query 62 GLVRDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERVESA--- 118
G+V D G+KMSKS N + ++I+ ++ L T +T +D+R E + S
Sbjct 543 GMVPDPFGKKMSKSHQNTLILQDLIQSEQTNSFDLHLFT-STNTDDIRNTDEMIRSNMVR 601
Query 119 ----------------------RNFANKLWNASRFMLMNLEGFDKT--FVPEASDYTLAD 154
+N K + +F N + V E++ + +
Sbjct 602 LRLCSICSFSDFTQPLNNTFNYQNILFKYYQIFKFQQKNTRDSVNSSHSVTESNSDPVVE 661
Query 155 KWILSRYAKTAISITENL 172
K I SR AK + + ENL
Sbjct 662 KLIRSRLAKLCVKLEENL 679
> dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=595
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 68 QGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGNDMRFYWERV 115
+G+KMSKS+GN I+P+E IEK+ D LR+ L+ P +D +Y ++V
Sbjct 343 KGKKMSKSIGNVINPLESIEKFTVDGLRYFLLRQGVPDSDCDYYDDKV 390
> eco:b0642 leuS, ECK0635, JW0637; leucyl-tRNA synthetase (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=860
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 43/81 (53%), Gaps = 5/81 (6%)
Query 48 EFGKDIPFKHVFIHGLVRDSQGRKMSKSLGNGIDPVEVIEKYGADTLRFMLITGNTPGND 107
E G+ + K H LV KMSKS NGIDP ++E+YGADT+R ++ + D
Sbjct 597 EKGRIVKAKDAAGHELVYTGMS-KMSKSKNNGIDPQVMVERYGADTVRLFMMFASPA--D 653
Query 108 MRFYWER--VESARNFANKLW 126
M W+ VE A F ++W
Sbjct 654 MTLEWQESGVEGANRFLKRVW 674
Lambda K H
0.323 0.138 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4406352944
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40