bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7884_orf1
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
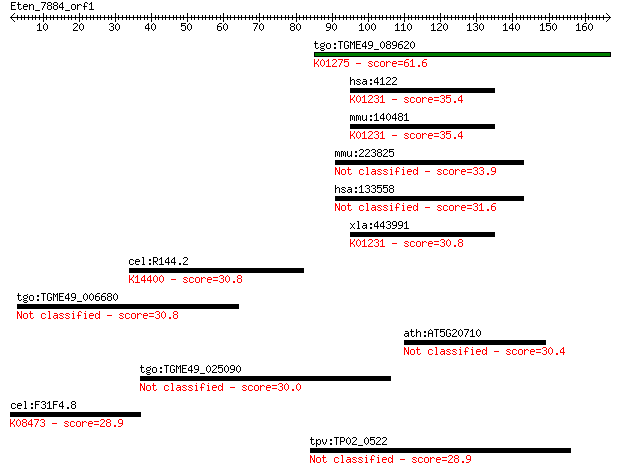
tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin ... 61.6 1e-09
hsa:4122 MAN2A2, MANA2X; mannosidase, alpha, class 2A, member ... 35.4 0.095
mmu:140481 Man2a2, 1700052O22Rik, 4931438M07Rik, AI480988, MX;... 35.4 0.097
mmu:223825 Heatr7b2, 4930455B06Rik, FLJ40243; XVHEAT repeat fa... 33.9 0.26
hsa:133558 HEATR7B2, DKFZp781F0822, FLJ40243; HEAT repeat fami... 31.6 1.4
xla:443991 man2a2, MGC80473; mannosidase, alpha, class 2A, mem... 30.8 2.2
cel:R144.2 hypothetical protein; K14400 pre-mRNA cleavage comp... 30.8 2.2
tgo:TGME49_006680 hypothetical protein 30.8 2.3
ath:AT5G20710 BGAL7; BGAL7 (beta-galactosidase 7); beta-galact... 30.4 2.9
tgo:TGME49_025090 hypothetical protein 30.0 3.7
cel:F31F4.8 srj-6; Serpentine Receptor, class J family member ... 28.9 8.2
tpv:TP02_0522 hypothetical protein 28.9 8.2
> tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin
C [EC:3.4.14.1]
Length=733
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 52/90 (57%), Gaps = 8/90 (8%)
Query 85 QELSYFLSEDFVLQHN-SKKSSWIAAVNSSFANQNKNFLNSLLKQFQ-SSPRLLGSSEGG 142
++ + ++S+ FV HN S SSW A VNS FAN N+ L+ +K F R G S G
Sbjct 250 KDSTSYISQSFVDTHNASPASSWRAGVNSVFANMNRRDLSRFVKDFGFKKMRPAGDSADG 309
Query 143 PVFLEEAAE------DAASTQAFACPCKEG 166
F++ + + DAASTQ +ACPCK+G
Sbjct 310 LSFMQSSTDISTADGDAASTQVYACPCKKG 339
> hsa:4122 MAN2A2, MANA2X; mannosidase, alpha, class 2A, member
2 (EC:3.2.1.114); K01231 alpha-mannosidase II [EC:3.2.1.114]
Length=1150
Score = 35.4 bits (80), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 95 FVLQHNSKKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPR 134
FV+ H+ WI + + Q ++ LNS++ + Q PR
Sbjct 169 FVVPHSHNDPGWIKTFDKYYTEQTQHILNSMVSKLQEDPR 208
> mmu:140481 Man2a2, 1700052O22Rik, 4931438M07Rik, AI480988, MX;
mannosidase 2, alpha 2; K01231 alpha-mannosidase II [EC:3.2.1.114]
Length=1152
Score = 35.4 bits (80), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 95 FVLQHNSKKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPR 134
FV+ H+ WI + + Q ++ LNS++ + Q PR
Sbjct 169 FVVPHSHNDPGWIKTFDKYYTEQTQHILNSMVSKLQEDPR 208
> mmu:223825 Heatr7b2, 4930455B06Rik, FLJ40243; XVHEAT repeat
family member 7B2
Length=1581
Score = 33.9 bits (76), Expect = 0.26, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 7/59 (11%)
Query 91 LSEDFVLQHNS-------KKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPRLLGSSEGG 142
LS DF LQ S KK W A + + Q+K+F++S + +F +P LLG G
Sbjct 590 LSRDFSLQMGSYSNSSMEKKFLWKALGTTLASCQDKDFVSSQINEFLVTPSLLGDHRQG 648
> hsa:133558 HEATR7B2, DKFZp781F0822, FLJ40243; HEAT repeat family
member 7B2
Length=1585
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 7/59 (11%)
Query 91 LSEDFVLQHNS-------KKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPRLLGSSEGG 142
L++DF Q S KK W A + Q+ +F+NS +K+F ++P LG G
Sbjct 594 LTQDFKQQMGSYSNNSTEKKFLWKALGTTLACCQDSDFVNSQIKEFLTAPNQLGDQRQG 652
> xla:443991 man2a2, MGC80473; mannosidase, alpha, class 2A, member
2 (EC:3.2.1.114); K01231 alpha-mannosidase II [EC:3.2.1.114]
Length=1150
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 11/40 (27%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 95 FVLQHNSKKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPR 134
FV+ H+ WI + + +Q ++ LN++L + PR
Sbjct 169 FVVPHSHNDPGWIKTFDKYYYDQTQHILNNMLVKLHEDPR 208
> cel:R144.2 hypothetical protein; K14400 pre-mRNA cleavage complex
2 protein Pcf11
Length=823
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 34 LRDSPAASAPSISRLGPVAFRPREFTFLNLTFSAFPFLFNYSPGPAAA 81
LRD P APS SR A P T N F F+ + +PG A+A
Sbjct 149 LRDDPQVMAPSQSRPAGNATSPAASTSTNRVFVNPKFIGSSTPGAASA 196
> tgo:TGME49_006680 hypothetical protein
Length=176
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 0/61 (0%)
Query 3 AASTCPFLFFSFPFLFFSFSFRFFSFFSPIQLRDSPAASAPSISRLGPVAFRPREFTFLN 62
AA+ CPF F+F SFF DSPA + +++ +G VA + ++
Sbjct 115 AAAACPFFKSGEERQIFAFDDLLGSFFHQRGQNDSPAKQSLNMTHVGKVAVSEEQRLEVD 174
Query 63 L 63
L
Sbjct 175 L 175
> ath:AT5G20710 BGAL7; BGAL7 (beta-galactosidase 7); beta-galactosidase
Length=826
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query 110 VNSSFANQNKNFLNSLLKQFQSSPRLLGSSEGGPVFLEE 148
VN SF N+ +NF ++K + L +S+GGP+ L +
Sbjct 144 VNPSFMNEMQNFTTKIVKMMKEEK--LFASQGGPIILAQ 180
> tgo:TGME49_025090 hypothetical protein
Length=2476
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query 37 SPAASAPSISRLGPVAFRPREFTFLNLT-FSAFPFLFNYSPGPAAAGG-AQELSYFLSED 94
SPA++AP + GP A P L LT +A L P P + GG Q S FLSE+
Sbjct 514 SPASAAPGVRTPGPAAAAP----VLVLTPRAACDALSRRGPAPLSGGGDRQGNSSFLSEE 569
Query 95 --FVLQHNSKKSS 105
H+ KK S
Sbjct 570 GRHFAAHDRKKHS 582
> cel:F31F4.8 srj-6; Serpentine Receptor, class J family member
(srj-6); K08473 nematode chemoreceptor
Length=331
Score = 28.9 bits (63), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query 1 FAAASTCPFLFFSFPFLFFSFSFRFFSFFSPIQLRD 36
+A +S C F+ S+ L F +R+F F P L D
Sbjct 88 YAISSRCTFIAISYALLIIHFVYRYFILFHP-HLVD 122
> tpv:TP02_0522 hypothetical protein
Length=1236
Score = 28.9 bits (63), Expect = 8.2, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 31/73 (42%), Gaps = 2/73 (2%)
Query 84 AQELSYFLSEDFV-LQHNSKKSSWIAAVNSSFANQNKNFLNSLLKQFQSSPRLLGSSEGG 142
EL F S D V L +N WI + +QN N L +LK F S L + G
Sbjct 121 GSELLQFESIDLVHLVNNGYPPKWINIYSKGIGSQNANLLGGILKFFSDSTSSL-TDYFG 179
Query 143 PVFLEEAAEDAAS 155
+ + +AE +
Sbjct 180 RILISASAEQVQT 192
Lambda K H
0.324 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40