bitscore colors: <40, 40-50 , 50-80, 80-200, >200
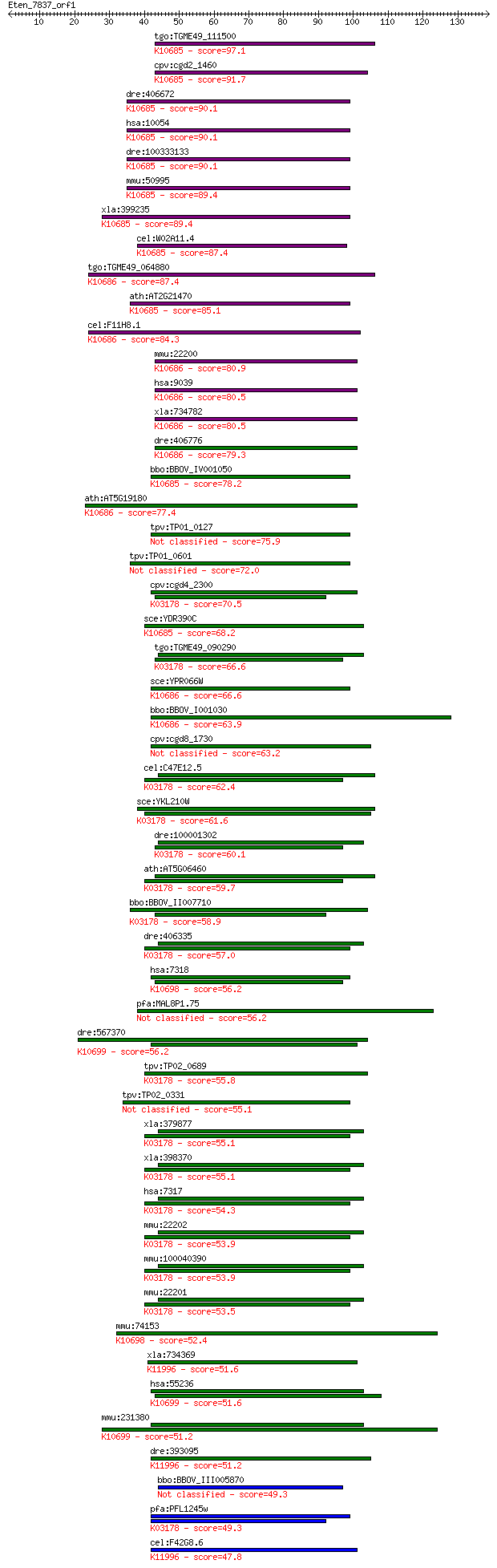
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7837_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.... 97.1 1e-20
cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiq... 91.7 6e-19
dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354; u... 90.1 2e-18
hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier a... 90.1 2e-18
dre:100333133 ubiquitin-like modifier activating enzyme 2-like... 90.1 2e-18
mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubi... 89.4 3e-18
xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-... 89.4 3e-18
cel:W02A11.4 uba-2; UBA (human ubiquitin) related family membe... 87.4 1e-17
tgo:TGME49_064880 ubiquitin-activating enzyme, putative ; K106... 87.4 1e-17
ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO acti... 85.1 6e-17
cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family m... 84.3 9e-17
mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539, U... 80.9 1e-15
hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin... 80.5 1e-15
xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier act... 80.5 2e-15
dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubi... 79.3 3e-15
bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K106... 78.2 8e-15
ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 acti... 77.4 1e-14
tpv:TP01_0127 ubiquitin-protein ligase 75.9 4e-14
tpv:TP01_0601 hypothetical protein 72.0 5e-13
cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178 ub... 70.5 2e-12
sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterod... 68.2 7e-12
tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ; K... 66.6 2e-11
sce:YPR066W UBA3; Protein that acts together with Ula1p to act... 66.6 2e-11
bbo:BBOV_I001030 16.m00748; ThiF family domain containing prot... 63.9 1e-10
cpv:cgd8_1730 Uba3p like ubiquitin activating enzyme E1 63.2 2e-10
cel:C47E12.5 uba-1; UBA (human ubiquitin) related family membe... 62.4 4e-10
sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme E1... 61.6 8e-10
dre:100001302 ubiquitin-like modifier-activating enzyme 1-like... 60.1 2e-09
ath:AT5G06460 ATUBA2; ATUBA2; ubiquitin activating enzyme/ ubi... 59.7 3e-09
bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1; K... 58.9 4e-09
dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:... 57.0 2e-08
hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-lik... 56.2 3e-08
pfa:MAL8P1.75 ubiquitin-activating enzyme, putative 56.2 3e-08
dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquit... 56.2 3e-08
tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activ... 55.8 4e-08
tpv:TP02_0331 ubiquitin activating enzyme, putatuve 55.1 7e-08
xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1, p... 55.1 7e-08
xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1, p... 55.1 7e-08
hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,... 54.3 1e-07
mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;... 53.9 1e-07
mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-li... 53.9 1e-07
mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-l... 53.5 2e-07
mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier ... 52.4 5e-07
xla:734369 mocs3, MGC84877, uba4; molybdenum cofactor synthesi... 51.6 7e-07
hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiq... 51.6 8e-07
mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799... 51.2 9e-07
dre:393095 mocs3, MGC55696, zgc:55696; molybdenum cofactor syn... 51.2 1e-06
bbo:BBOV_III005870 17.m07520; ThiF family protein 49.3 4e-06
pfa:PFL1245w ubiquitin-activating enzyme E1, putative; K03178 ... 49.3 4e-06
cel:F42G8.6 hypothetical protein; K11996 adenylyltransferase a... 47.8 1e-05
> tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.2.1.70);
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=730
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/63 (71%), Positives = 53/63 (84%), Gaps = 0/63 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAAA 102
VLVVGAGGIGCE+ K L+L GF R+ V+DLD+ID++NLNRQFFFR HVG SKA VLAAA
Sbjct 39 VLVVGAGGIGCEVCKDLLLSGFRRLCVVDLDTIDVSNLNRQFFFRNAHVGLSKAFVLAAA 98
Query 103 AAA 105
+A
Sbjct 99 CSA 101
> cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=637
Score = 91.7 bits (226), Expect = 6e-19, Method: Composition-based stats.
Identities = 40/61 (65%), Positives = 51/61 (83%), Gaps = 0/61 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAAA 102
+LVVGAGGIGCELVK L+L GF I ++D+D IDI+NLNRQFFFR++HVG +K+ V+A
Sbjct 24 ILVVGAGGIGCELVKDLILSGFSNITIIDMDGIDISNLNRQFFFRRKHVGMNKSTVVALE 83
Query 103 A 103
A
Sbjct 84 A 84
> dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354;
ubiquitin-like modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=640
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 42/64 (65%), Positives = 53/64 (82%), Gaps = 0/64 (0%)
Query 35 SSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSS 94
+ S+ S VLVVGAGGIGCEL+K+LVL GF I V+DLD+ID++NLNRQF F+++HVG S
Sbjct 13 ADSLSSCRVLVVGAGGIGCELLKNLVLTGFKNIEVIDLDTIDVSNLNRQFLFQKKHVGKS 72
Query 95 KAAV 98
KA V
Sbjct 73 KAQV 76
> hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier
activating enzyme 2; K10685 ubiquitin-like 1-activating enzyme
E1 B [EC:6.3.2.19]
Length=640
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 41/64 (64%), Positives = 53/64 (82%), Gaps = 0/64 (0%)
Query 35 SSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSS 94
+ +V G VLVVGAGGIGCEL+K+LVL GF I ++DLD+ID++NLNRQF F+++HVG S
Sbjct 12 AEAVAGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNRQFLFQKKHVGRS 71
Query 95 KAAV 98
KA V
Sbjct 72 KAQV 75
> dre:100333133 ubiquitin-like modifier activating enzyme 2-like;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=642
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 42/64 (65%), Positives = 53/64 (82%), Gaps = 0/64 (0%)
Query 35 SSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSS 94
+ S+ S VLVVGAGGIGCEL+K+LVL GF I V+DLD+ID++NLNRQF F+++HVG S
Sbjct 13 ADSLSSCRVLVVGAGGIGCELLKNLVLTGFKNIEVIDLDTIDVSNLNRQFLFQKKHVGKS 72
Query 95 KAAV 98
KA V
Sbjct 73 KAQV 76
> mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubiquitin-like
modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=638
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 41/64 (64%), Positives = 53/64 (82%), Gaps = 0/64 (0%)
Query 35 SSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSS 94
+ +V G VLVVGAGGIGCEL+K+LVL GF I ++DLD+ID++NLNRQF F+++HVG S
Sbjct 12 AEAVSGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNRQFLFQKKHVGRS 71
Query 95 KAAV 98
KA V
Sbjct 72 KAQV 75
> xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-b,
uble1b-B; ubiquitin-like modifier activating enzyme 2;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=641
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 41/71 (57%), Positives = 55/71 (77%), Gaps = 0/71 (0%)
Query 28 GATSTASSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFR 87
GA + +V + +LVVGAGGIGCEL+K+LVL GF + V+DLD+ID++NLNRQF F+
Sbjct 5 GALPKEVAEAVSASRLLVVGAGGIGCELLKNLVLTGFTNLDVIDLDTIDVSNLNRQFLFQ 64
Query 88 QQHVGSSKAAV 98
++HVG SKA V
Sbjct 65 KKHVGRSKAQV 75
> cel:W02A11.4 uba-2; UBA (human ubiquitin) related family member
(uba-2); K10685 ubiquitin-like 1-activating enzyme E1 B
[EC:6.3.2.19]
Length=582
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 37/60 (61%), Positives = 50/60 (83%), Gaps = 0/60 (0%)
Query 38 VCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAA 97
+ +LV+GAGGIGCEL+K+L + GF ++ V+DLD+IDI+NLNRQF FR++HV SSKAA
Sbjct 11 IVQSKILVIGAGGIGCELLKNLAVTGFRKVHVIDLDTIDISNLNRQFLFRKEHVSSSKAA 70
> tgo:TGME49_064880 ubiquitin-activating enzyme, putative ; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=668
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 54/82 (65%), Gaps = 4/82 (4%)
Query 24 FSPGGATSTASSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQ 83
F PG T + VLVVGAGG+GCE++K L L GF R+ V+D+D+I +TNL+RQ
Sbjct 47 FEPGAETIERLRDT----HVLVVGAGGLGCEVLKCLCLSGFRRLDVIDMDTIHVTNLHRQ 102
Query 84 FFFRQQHVGSSKAAVLAAAAAA 105
F FR++HVG KA V A A A
Sbjct 103 FLFREKHVGRPKAQVAAEALNA 124
> ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO activating
enzyme; K10685 ubiquitin-like 1-activating enzyme E1
B [EC:6.3.2.19]
Length=625
Score = 85.1 bits (209), Expect = 6e-17, Method: Composition-based stats.
Identities = 37/63 (58%), Positives = 50/63 (79%), Gaps = 0/63 (0%)
Query 36 SSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSK 95
S++ VL+VGAGGIGCEL+K+L L GF I ++D+D+I+++NLNRQF FR+ HVG SK
Sbjct 8 SAIKGAKVLMVGAGGIGCELLKTLALSGFEDIHIIDMDTIEVSNLNRQFLFRRSHVGQSK 67
Query 96 AAV 98
A V
Sbjct 68 AKV 70
> cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family
member (rfl-1); K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=430
Score = 84.3 bits (207), Expect = 9e-17, Method: Composition-based stats.
Identities = 41/78 (52%), Positives = 54/78 (69%), Gaps = 4/78 (5%)
Query 24 FSPGGATSTASSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQ 83
F PG A ++ +LV+GAGG+GCEL+K+L L GF I V+D+D+ID++NLNRQ
Sbjct 30 FVPGPENFEALQNT----KILVIGAGGLGCELLKNLALSGFRTIEVIDMDTIDVSNLNRQ 85
Query 84 FFFRQQHVGSSKAAVLAA 101
F FR+ VG SKA V AA
Sbjct 86 FLFRESDVGKSKAEVAAA 103
> mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539,
Ube1c; ubiquitin-like modifier activating enzyme 3; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=448
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 36/58 (62%), Positives = 47/58 (81%), Gaps = 0/58 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
VLV+GAGG+GCEL+K+L L GF +I V+D+D+ID++NLNRQF FR + VG KA V A
Sbjct 58 VLVIGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDVGRPKAEVAA 115
> hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin-like
modifier activating enzyme 3; K10686 ubiquitin-activating
enzyme E1 C [EC:6.3.2.19]
Length=463
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 35/58 (60%), Positives = 47/58 (81%), Gaps = 0/58 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
VLV+GAGG+GCEL+K+L L GF +I V+D+D+ID++NLNRQF FR + +G KA V A
Sbjct 72 VLVIGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDIGRPKAEVAA 129
> xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier activating
enzyme 3; K10686 ubiquitin-activating enzyme E1 C
[EC:6.3.2.19]
Length=461
Score = 80.5 bits (197), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/58 (62%), Positives = 47/58 (81%), Gaps = 0/58 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
+LVVGAGG+GCEL+K+L L GF +I V+D+D+ID++NLNRQF FR + VG KA V A
Sbjct 71 LLVVGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDVGRPKAEVAA 128
> dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubiquitin-like
modifier activating enzyme 3 (EC:6.3.2.-); K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=462
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/58 (60%), Positives = 45/58 (77%), Gaps = 0/58 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
+LV+GAGG+GCEL+K L L GF I V+D+D+ID++NLNRQF FR + VG KA V A
Sbjct 71 ILVIGAGGLGCELLKDLALSGFRHIHVVDMDTIDVSNLNRQFLFRPKDVGRPKAEVAA 128
> bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=630
Score = 78.2 bits (191), Expect = 8e-15, Method: Composition-based stats.
Identities = 34/57 (59%), Positives = 47/57 (82%), Gaps = 0/57 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
S+LVVGAGGIGCEL+K+LVL G ++++D+D+ID++NLNRQF +R + VG KA V
Sbjct 47 SLLVVGAGGIGCELIKNLVLCGVRNLVIVDIDTIDVSNLNRQFLYRAEDVGRYKAEV 103
> ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 activating
enzyme/ protein heterodimerization/ small protein activating
enzyme; K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=454
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 39/78 (50%), Positives = 49/78 (62%), Gaps = 3/78 (3%)
Query 23 GFSPGGATSTASSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNR 82
GF PG V +LV+GAGG+GCEL+K L L GF + V+D+D I++TNLNR
Sbjct 32 GFVPGPGLRDDIRDYV---RILVIGAGGLGCELLKDLALSGFRNLEVIDMDRIEVTNLNR 88
Query 83 QFFFRQQHVGSSKAAVLA 100
QF FR + VG KA V A
Sbjct 89 QFLFRIEDVGKPKAEVAA 106
> tpv:TP01_0127 ubiquitin-protein ligase
Length=543
Score = 75.9 bits (185), Expect = 4e-14, Method: Composition-based stats.
Identities = 31/57 (54%), Positives = 46/57 (80%), Gaps = 0/57 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
SVL+VGAGGIGCEL+K+L+L G ++ ++D+D++D++NLNRQF + +HV KA V
Sbjct 28 SVLLVGAGGIGCELIKTLLLTGVKKLTIVDMDTVDVSNLNRQFLYLPEHVNKYKAEV 84
> tpv:TP01_0601 hypothetical protein
Length=343
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 31/63 (49%), Positives = 46/63 (73%), Gaps = 0/63 (0%)
Query 36 SSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSK 95
++V +LVVG+GG+GCEL+KSLVL GF I ++D D + ++NLNRQF F++ VG K
Sbjct 2 NNVLKSRILVVGSGGLGCELLKSLVLNGFENISIVDFDKVVLSNLNRQFLFQKNDVGKFK 61
Query 96 AAV 98
+ +
Sbjct 62 SQI 64
> cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178
ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1067
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 30/64 (46%), Positives = 46/64 (71%), Gaps = 5/64 (7%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
++ +VGAG +GCE +KS+ L G + + D+D+I+++NLNRQF FRQ+HVGS K+
Sbjct 457 NIFIVGAGALGCEFLKSMALLGVGCGPNGTVTITDMDNIEVSNLNRQFLFRQEHVGSPKS 516
Query 97 AVLA 100
A+ A
Sbjct 517 AIAA 520
Score = 31.6 bits (70), Expect = 0.82, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHV 91
VL+VG G+G E+ K+++L G I ++D + +++ F+ + V
Sbjct 39 VLIVGLRGLGVEIAKNIILAGPKSITLVDDEICSFSDMGANFYITENDV 87
> sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterodimer
with Aos1p to activate Smt3p (SUMO) before its conjugation
to proteins (sumoylation), which may play a role in protein
targeting; essential for viability; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=636
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 31/63 (49%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVL 99
S L+VGAGGIG EL+K ++L F I ++DLD+ID++NLNRQF FRQ+ + K+
Sbjct 21 SSRCLLVGAGGIGSELLKDIILMEFGEIHIVDLDTIDLSNLNRQFLFRQKDIKQPKSTTA 80
Query 100 AAA 102
A
Sbjct 81 VKA 83
> tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1091
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 45/66 (68%), Gaps = 7/66 (10%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-------RILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
VVGAG +GCEL+KSL L G ++ V D+D I+++NLNRQF FR++HVG +K+
Sbjct 470 FVVGAGALGCELLKSLALMGCGCGPEKEGKVTVTDMDRIEVSNLNRQFLFRREHVGKAKS 529
Query 97 AVLAAA 102
AA+
Sbjct 530 VTAAAS 535
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHV--GSSKA 96
VL+ G G+G E K+L+L G +++ D ++ +L F ++HV G S+A
Sbjct 39 VLISGMRGVGAECAKNLILAGPNTVVLHDPAPCEMRDLGSNFCLTEEHVKKGVSRA 94
> sce:YPR066W UBA3; Protein that acts together with Ula1p to activate
Rub1p before its conjugation to proteins (neddylation),
which may play a role in protein degradation; GFP-fusion
protein localizes to the cytoplasm in a punctate pattern (EC:6.3.2.-);
K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=299
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 44/58 (75%), Gaps = 1/58 (1%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYR-ILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+LV+GAGG+GCE++K+L + F + + ++D+D+I++TNLNRQF F + +G KA V
Sbjct 4 KILVLGAGGLGCEILKNLTMLSFVKQVHIVDIDTIELTNLNRQFLFCDKDIGKPKAQV 61
> bbo:BBOV_I001030 16.m00748; ThiF family domain containing protein;
K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=375
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 50/86 (58%), Gaps = 1/86 (1%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAA 101
+V+V+GAGG+GCE++K++VL G I ++D D I+I N+ RQF ++ VG K A++AA
Sbjct 6 NVIVIGAGGLGCEVIKNIVLLGSRNITIVDPDIIEIHNITRQFLYKVDDVGKYK-AIVAA 64
Query 102 AAAADPAAAAAAAAAAPADAETPAAA 127
+ + A E P +
Sbjct 65 ERIKECNSNIKVEAITKRAQELPISV 90
> cpv:cgd8_1730 Uba3p like ubiquitin activating enzyme E1
Length=346
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 42/63 (66%), Gaps = 0/63 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAA 101
VL+VG GGIG E+++ L+ GF RI ++D D ++++N++RQ FF G SK VLAA
Sbjct 53 KVLLVGVGGIGTEILRCLIFSGFRRIDIVDYDYVEVSNISRQLFFNLGDEGKSKVHVLAA 112
Query 102 AAA 104
A
Sbjct 113 NAT 115
> cel:C47E12.5 uba-1; UBA (human ubiquitin) related family member
(uba-1); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1113
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 32/67 (47%), Positives = 42/67 (62%), Gaps = 5/67 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
VVGAG IGCEL+K+L + G I + D+D I+I+NLNRQF FR++ VG K+
Sbjct 521 FVVGAGAIGCELLKNLSMMGVACGEGGLIKITDMDQIEISNLNRQFLFRRRDVGGKKSEC 580
Query 99 LAAAAAA 105
A A A
Sbjct 581 AARAVTA 587
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
+ SVL+ G G +G E+ K+L+L G + + D ++L+ Q++ R VG ++A
Sbjct 125 TASVLISGLGSVGVEIAKNLILGGVRHVTIHDTKLAKWSDLSAQYYLRDADVGHNRA 181
> sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1024
Score = 61.6 bits (148), Expect = 8e-10, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 45/73 (61%), Gaps = 5/73 (6%)
Query 38 VCSGSVLVVGAGGIGCELVKSLVLRGFYR-----ILVLDLDSIDITNLNRQFFFRQQHVG 92
+ + V +VG+G IGCE++K+ L G I+V D DSI+ +NLNRQF FR + VG
Sbjct 432 IANSKVFLVGSGAIGCEMLKNWALLGLGSGSDGYIVVTDNDSIEKSNLNRQFLFRPKDVG 491
Query 93 SSKAAVLAAAAAA 105
+K+ V A A A
Sbjct 492 KNKSEVAAEAVCA 504
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 38/65 (58%), Gaps = 0/65 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVL 99
+ +VL++G G+G E+ K++VL G + V D + + + +L+ QFF ++ +G + V
Sbjct 36 TSNVLILGLKGLGVEIAKNVVLAGVKSMTVFDPEPVQLADLSTQFFLTEKDIGQKRGDVT 95
Query 100 AAAAA 104
A A
Sbjct 96 RAKLA 100
> dre:100001302 ubiquitin-like modifier-activating enzyme 1-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1016
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/64 (48%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ L G I V D+DSI+ +NLNRQF FR Q +G K+
Sbjct 430 FLVGAGAIGCELLKNFALIGLGAGEGGSITVTDMDSIERSNLNRQFLFRSQDIGRPKSEA 489
Query 99 LAAA 102
A A
Sbjct 490 AAEA 493
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
VL+ G G+G E+ K+++L G + + D ++ +L+ QF+ ++ +G ++A
Sbjct 31 VLIAGMRGLGVEIAKNVILAGVRTVTIQDEGVVEWRDLSSQFYLKEADLGQNRA 84
> ath:AT5G06460 ATUBA2; ATUBA2; ubiquitin activating enzyme/ ubiquitin-protein
ligase; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1077
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 31/68 (45%), Positives = 43/68 (63%), Gaps = 5/68 (7%)
Query 43 VLVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAA 97
V VVGAG +GCE +K+L L G ++ V D D I+ +NL+RQF FR ++G +K+
Sbjct 492 VFVVGAGALGCEFLKNLALMGVSCGTQGKLTVTDDDVIEKSNLSRQFLFRDWNIGQAKST 551
Query 98 VLAAAAAA 105
V A AAA
Sbjct 552 VAATAAAG 559
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
+ +VL+ G G+G E+ K+++L G + + D + +++ +L+ F F ++ +G ++A
Sbjct 92 ASNVLISGMQGLGVEIAKNIILAGVKSVTLHDENVVELWDLSSNFVFTEEDIGKNRA 148
> bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1007
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 44/73 (60%), Gaps = 5/73 (6%)
Query 36 SSVCSGSVLVVGAGGIGCELVKSLVLRGFYR-----ILVLDLDSIDITNLNRQFFFRQQH 90
S + S + VG+G +GCE +K L G + + D D I+++N++RQF FR++H
Sbjct 416 SKIQSAKIFTVGSGALGCEFMKHFALLGCGTQNGGIVKITDNDRIEVSNISRQFLFRKKH 475
Query 91 VGSSKAAVLAAAA 103
VG SK+ V A +A
Sbjct 476 VGMSKSKVAAISA 488
Score = 35.0 bits (79), Expect = 0.063, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHV 91
VL++G G+G E+ K+L L G I + D + ++ +L FF R V
Sbjct 35 VLILGMKGVGVEIAKNLALMGVEAICITDDNIVERRDLGVNFFIRSSDV 83
> dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:fj14g11,
zgc:66143; ubiquitin-like modifier activating enzyme
1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/63 (44%), Positives = 39/63 (61%), Gaps = 4/63 (6%)
Query 44 LVVGAGGIGCELVKSLVLRGFY----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVL 99
+VGAG IGCEL+K+ + G ++V D+D+I+ +NLNRQF FR V K+
Sbjct 473 FLVGAGAIGCELLKNFAMMGLASGEGEVIVTDMDTIEKSNLNRQFLFRPWDVTKMKSETA 532
Query 100 AAA 102
AAA
Sbjct 533 AAA 535
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
S +VL+ G G+G E+ K+++L G + + D + +L+ QF+ R++ +G ++A V
Sbjct 72 SSNVLISGLRGLGVEIAKNVILGGVKSVTLHDQGVAEWKDLSSQFYLREEDLGKNRADV 130
> hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-like
modifier activating enzyme 7; K10698 ubiquitin-activating
enzyme E1-like [EC:6.3.2.19]
Length=1012
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 38/62 (61%), Gaps = 5/62 (8%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYR-----ILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
L+VGAG IGCEL+K L G + V+D+D I+ +NL+RQF FR Q VG KA
Sbjct 434 HYLLVGAGAIGCELLKVFALVGLGAGNSGGLTVVDMDHIERSNLSRQFLFRSQDVGRPKA 493
Query 97 AV 98
V
Sbjct 494 EV 495
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
VLV G G+G E+ K+LVL G + + D ++L QF +Q + S+A
Sbjct 35 VLVSGLQGLGAEVAKNLVLMGVGSLTLHDPHPTCWSDLAAQFLLSEQDLERSRA 88
> pfa:MAL8P1.75 ubiquitin-activating enzyme, putative
Length=389
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 43/90 (47%), Gaps = 5/90 (5%)
Query 38 VCSGS-----VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVG 92
+C+ S +LVVG GG+G E++K+L+ I ++D D ++++NL RQ FF +G
Sbjct 5 ICTKSFERLNILVVGCGGLGNEVIKNLIFLHIKNICIVDYDIVEVSNLQRQLFFSHDDIG 64
Query 93 SSKAAVLAAAAAADPAAAAAAAAAAPADAE 122
K V++ + E
Sbjct 65 KYKVDVISYKIKETYMHENICIKSYKNHIE 94
> dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1060
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 36/97 (37%), Positives = 47/97 (48%), Gaps = 14/97 (14%)
Query 21 AAGFSPGG----ATSTASSSSVC----SGSVLVVGAGGIGCELVKSLVLRG------FYR 66
A FSP G A S+C V +VG G IGCE++K+L L G
Sbjct 434 AEEFSPRGDRYDALRACIGQSLCLKLHKFQVFMVGCGAIGCEMLKNLALLGVGLSRFLGE 493
Query 67 ILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAAAA 103
I + D D I+ +NLNRQF FR H+ K+ A A+
Sbjct 494 ICITDPDLIEKSNLNRQFLFRPHHIQKPKSTTAAEAS 530
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
+V V G G +G E+ K++VL G + + D ++ +L FF R++ V + K V A
Sbjct 64 TVFVSGMGALGVEIAKNIVLAGVKAVTLHDSKRCEVWDLGTNFFIREEDVNNQKKRVEA 122
> tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=999
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 43/69 (62%), Gaps = 5/69 (7%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYR-----ILVLDLDSIDITNLNRQFFFRQQHVGSS 94
+ + +VGAG +GCE +K+ L G + + D D I+++N++RQF FR +HVG +
Sbjct 416 NSKIFIVGAGALGCEFLKNFALLGCGSQPDGLLTITDNDRIEVSNISRQFLFRTRHVGLA 475
Query 95 KAAVLAAAA 103
K++V +A
Sbjct 476 KSSVACESA 484
> tpv:TP02_0331 ubiquitin activating enzyme, putatuve
Length=1126
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 34 SSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGS 93
S V S LVVGAG +GC+ +K L G + V D D++D++NL RQ F VG
Sbjct 483 SMEQVAKMSFLVVGAGALGCDYLKLLAEMGVSDVTVFDNDTVDVSNLTRQVLFTINDVGK 542
Query 94 SKAAV 98
KA V
Sbjct 543 PKAQV 547
> xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1a, ube1, ube1x; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1059
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G I V D+D+I+ +NLNRQF FR V K+
Sbjct 473 FLVGAGAIGCELLKNFAMIGLAAGDGGEITVTDMDTIEKSNLNRQFLFRPWDVTKMKSDT 532
Query 99 LAAA 102
AAA
Sbjct 533 AAAA 536
Score = 44.7 bits (104), Expect = 9e-05, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ +VL+ G G+G E+ K+++L G + + D + D +L+ QF+ R+ +G ++A V
Sbjct 71 NSNVLISGMSGLGVEIAKNIILAGVKSVTIHDQHNTDWADLSSQFYLRESDIGKNRAEV 129
> xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1, uba1a, uba1b, ube, ube1, ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1060
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G I V D+D+I+ +NLNRQF FR V K+
Sbjct 474 FLVGAGAIGCELLKNFAMIGLAAGEGGEITVTDMDTIEKSNLNRQFLFRPWDVTKMKSDT 533
Query 99 LAAA 102
AAA
Sbjct 534 AAAA 537
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ +VL+ G G+G E+ K+++L G + + D + + T+L+ QF+ R+ +G ++A V
Sbjct 72 NSNVLISGMSGLGVEIAKNIILAGVKSVTIHDQHNTEWTDLSSQFYLRESDIGKNRAEV 130
> hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,
SMAX2, UBA1A, UBE1, UBE1X; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1058
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G I+V D+D+I+ +NLNRQF FR V K+
Sbjct 472 FLVGAGAIGCELLKNFAMIGLGCGEGGEIIVTDMDTIEKSNLNRQFLFRPWDVTKLKSDT 531
Query 99 LAAA 102
AAA
Sbjct 532 AAAA 535
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ SVLV G G+G E+ K+++L G + + D + +L+ QF+ R++ +G ++A V
Sbjct 72 TSSVLVSGLRGLGVEIAKNIILGGVKAVTLHDQGTAQWADLSSQFYLREEDIGKNRAEV 130
> mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;
ubiquitin-activating enzyme E1, Chr Y 1; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G I V D+D+I+ +NLNRQF FR + K+
Sbjct 471 FLVGAGAIGCELLKNFAMIGLGCGEDGEITVTDMDTIEKSNLNRQFLFRPWDITKLKSET 530
Query 99 LAAA 102
AAA
Sbjct 531 AAAA 534
Score = 38.1 bits (87), Expect = 0.009, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ SVL+ G G+G E+ K+++L G + + D +L+ QF R++ +G ++A +
Sbjct 71 ASSVLISGLQGLGVEIAKNIILGGVKAVTLHDQGIAQWADLSSQFCLREEDIGKNRAEI 129
> mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G I V D+D+I+ +NLNRQF FR + K+
Sbjct 471 FLVGAGAIGCELLKNFAMIGLGCGEDGEITVTDMDTIEKSNLNRQFLFRPWDITKLKSET 530
Query 99 LAAA 102
AAA
Sbjct 531 AAAA 534
Score = 38.1 bits (87), Expect = 0.009, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ SVL+ G G+G E+ K+++L G + + D +L+ QF R++ +G ++A +
Sbjct 71 ASSVLISGLQGLGVEIAKNIILGGVKAVTLHDQGIAQWADLSSQFCLREEDIGKNRAEI 129
> mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 44 LVVGAGGIGCELVKSLVLRGFY-----RILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+VGAG IGCEL+K+ + G ++V D+D+I+ +NLNRQF FR V K+
Sbjct 472 FLVGAGAIGCELLKNFAMIGLGCGEGGEVVVTDMDTIEKSNLNRQFLFRPWDVTKLKSDT 531
Query 99 LAAA 102
AAA
Sbjct 532 AAAA 535
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 40 SGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAV 98
+ SVLV G G+G E+ K+++L G + + D + +L+ QF+ R++ +G ++A V
Sbjct 72 TSSVLVSGLRGLGVEIAKNIILGGVKAVTLHDQGTTQWADLSSQFYLREEDIGKNRAEV 130
> mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier
activating enzyme 7; K10698 ubiquitin-activating enzyme E1-like
[EC:6.3.2.19]
Length=977
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 47/97 (48%), Gaps = 5/97 (5%)
Query 32 TASSSSVCSGSVLVVGAGGIGCELVKSLVLRGF-YR----ILVLDLDSIDITNLNRQFFF 86
T + L+VGAG IGCE++K L G R + V D+D I+ +NL+RQF F
Sbjct 411 TDLQEKLSDQHYLLVGAGAIGCEMLKVFALVGLGVRANGGVTVADMDYIERSNLSRQFLF 470
Query 87 RQQHVGSSKAAVLAAAAAADPAAAAAAAAAAPADAET 123
R + V KA V AAAA A P D T
Sbjct 471 RPKDVRRPKAEVAAAAAHRLNPDLRATPYTCPLDPTT 507
> xla:734369 mocs3, MGC84877, uba4; molybdenum cofactor synthesis
3 (EC:2.7.7.- 2.8.1.-); K11996 adenylyltransferase and sulfurtransferase
Length=451
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 41 GSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
SVLV+G GG+GC + + L G R+ +LD D ++++NL+RQ + +G SK+ +A
Sbjct 77 ASVLVIGCGGLGCPVAQYLAASGIGRLGLLDYDVVEMSNLHRQVLHGENRLGMSKSVSVA 136
> hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiquitin-like
modifier activating enzyme 6; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1052
Score = 51.6 bits (122), Expect = 8e-07, Method: Composition-based stats.
Identities = 27/67 (40%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYR------ILVLDLDSIDITNLNRQFFFRQQHVGSSK 95
++ +VG G IGCE++K+ L G I V D D I+ +NLNRQF FR H+ K
Sbjct 462 NIFLVGCGAIGCEMLKNFALLGVGTSKEKGMITVTDPDLIEKSNLNRQFLFRPHHIQKPK 521
Query 96 AAVLAAA 102
+ A A
Sbjct 522 SYTAADA 528
Score = 35.4 bits (80), Expect = 0.053, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 43 VLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSK---AAVL 99
V + G GG+G E+ K+LVL G + + D + +L FF + V + + AVL
Sbjct 64 VFLSGMGGLGLEIAKNLVLAGIKAVTIHDTEKCQAWDLGTNFFLSEDDVVNKRNRAEAVL 123
Query 100 AAAAAADP 107
A +P
Sbjct 124 KHIAELNP 131
> mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799,
E1-L2, Ube1l2; ubiquitin-like modifier activating enzyme
6; K10699 ubiquitin-activating enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1053
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYR------ILVLDLDSIDITNLNRQFFFRQQHVGSSK 95
++ +VG G IGCE++K+ L G + V D D I+ +NLNRQF FR H+ K
Sbjct 462 NIFLVGCGAIGCEMLKNFALLGVGTGREKGMVTVTDPDLIEKSNLNRQFLFRPHHIQKPK 521
Query 96 AAVLAAA 102
+ A A
Sbjct 522 SYTAAEA 528
Score = 37.4 bits (85), Expect = 0.015, Method: Composition-based stats.
Identities = 28/99 (28%), Positives = 45/99 (45%), Gaps = 7/99 (7%)
Query 28 GATSTASSSSVCSGSVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFR 87
G T+ + C V + G GG+G E+ K+LVL G + + D +L FF
Sbjct 52 GDTAMQKMAKSC---VFLSGMGGLGVEIAKNLVLAGIKALTIHDTKKCQAWDLGTNFFLC 108
Query 88 QQHVGSSK---AAVLAAAAAADPAAAAAAAAAAPADAET 123
+ V + + AVL A +P ++++AP D T
Sbjct 109 EDDVVNERNRAEAVLHRIAELNP-YVQVSSSSAPLDETT 146
> dre:393095 mocs3, MGC55696, zgc:55696; molybdenum cofactor synthesis
3 (EC:2.8.1.-); K11996 adenylyltransferase and sulfurtransferase
Length=459
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 28/63 (44%), Positives = 39/63 (61%), Gaps = 2/63 (3%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLAA 101
SVLVVG GG+GC L + L G R+ +LD D ++++NL+RQ + G KA L+A
Sbjct 84 SVLVVGCGGLGCPLAQYLAAAGIGRLGLLDYDVVELSNLHRQVLHTELTQGQPKA--LSA 141
Query 102 AAA 104
A A
Sbjct 142 AQA 144
> bbo:BBOV_III005870 17.m07520; ThiF family protein
Length=1009
Score = 49.3 bits (116), Expect = 4e-06, Method: Composition-based stats.
Identities = 23/53 (43%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 44 LVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKA 96
L+VG G +GCE +K L G + +D DS+D++NL RQ F VG +KA
Sbjct 393 LMVGVGALGCEYLKILEAMGVEHLTAMDNDSVDVSNLTRQSLFTDADVGLNKA 445
> pfa:PFL1245w ubiquitin-activating enzyme E1, putative; K03178
ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1140
Score = 49.3 bits (116), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 40/80 (50%), Gaps = 23/80 (28%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFY-----------------------RILVLDLDSIDIT 78
+V +VG+G +GCE K L ++ + D D+I+++
Sbjct 505 NVFLVGSGALGCEYAKLFSLLDMCTRNSEQNTNLNQNNIDNNLACCGKLTITDNDNIEVS 564
Query 79 NLNRQFFFRQQHVGSSKAAV 98
NLNRQF FR++HVG SK+ V
Sbjct 565 NLNRQFLFRREHVGKSKSLV 584
Score = 35.0 bits (79), Expect = 0.065, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHV 91
++L++ G+G E K+L+L G + + D D DI+++ F+ ++ V
Sbjct 67 NILIINVKGVGLECAKNLILSGPQSVCIYDNDICDISDIGVNFYINEKDV 116
> cel:F42G8.6 hypothetical protein; K11996 adenylyltransferase
and sulfurtransferase
Length=402
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 42 SVLVVGAGGIGCELVKSLVLRGFYRILVLDLDSIDITNLNRQFFFRQQHVGSSKAAVLA 100
+VL+VGAGG+GC + L G I ++D D I + NL+RQ +++ VG SKA LA
Sbjct 39 NVLIVGAGGLGCPVATYLGAAGIGTIGIVDYDHISLDNLHRQVAYKEDQVGKSKAQALA 97
Lambda K H
0.315 0.127 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40