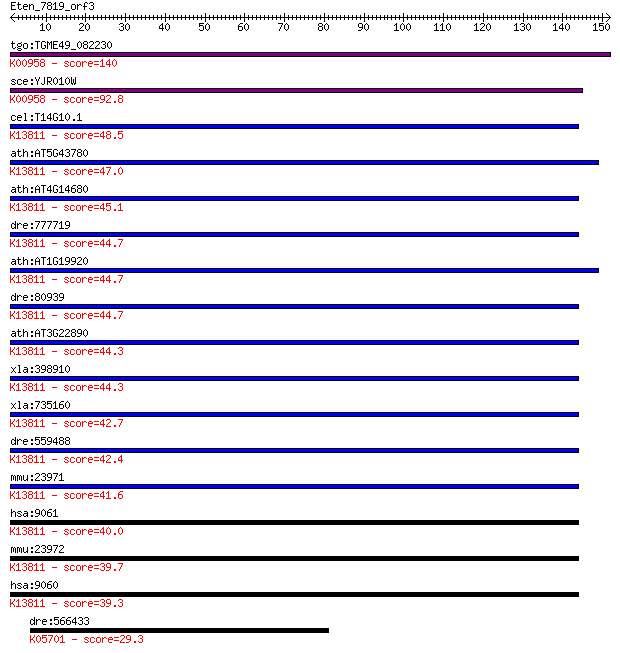
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7819_orf3
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate k... 140 2e-33
sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step ... 92.8 4e-19
cel:T14G10.1 pps-1; 3'-Phosphoadenosine 5'-Phosphosulfate Synt... 48.5 9e-06
ath:AT5G43780 APS4; APS4; sulfate adenylyltransferase (ATP); K... 47.0 2e-05
ath:AT4G14680 APS3; APS3; sulfate adenylyltransferase (ATP); K... 45.1 9e-05
dre:777719 papss2a, zgc:153748; 3'-phosphoadenosine 5'-phospho... 44.7 1e-04
ath:AT1G19920 APS2; APS2; sulfate adenylyltransferase (ATP); K... 44.7 1e-04
dre:80939 papss2b, Papss1, id:ibd2761, papss2, sb:cb868, wu:fb... 44.7 1e-04
ath:AT3G22890 APS1; APS1 (ATP SULFURYLASE 1); sulfate adenylyl... 44.3 1e-04
xla:398910 papss2, MGC68677; 3'-phosphoadenosine 5'-phosphosul... 44.3 1e-04
xla:735160 papss1, MGC114937, atpsk1, papss, sk1; 3'-phosphoad... 42.7 5e-04
dre:559488 papss1, wu:fc43e12, zgc:194984, zgc:194985; 3'-phos... 42.4 6e-04
mmu:23971 Papss1, AI325286, Asapk, SK1; 3'-phosphoadenosine 5'... 41.6 0.001
hsa:9061 PAPSS1, ATPSK1, PAPSS, SK1; 3'-phosphoadenosine 5'-ph... 40.0 0.003
mmu:23972 Papss2, 1810018P12Rik, AI159688, AtpsU2, Atpsk2, Sk2... 39.7 0.003
hsa:9060 PAPSS2, ATPSK2, SK2; 3'-phosphoadenosine 5'-phosphosu... 39.3 0.005
dre:566433 tight junction protein 1-like; K05701 tight junctio... 29.3 5.3
> tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate
kinase, putative (EC:2.7.7.4 2.7.1.25); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=607
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 71/151 (47%), Positives = 83/151 (54%), Gaps = 43/151 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
HRSHYEL K AL + K+P LLLTP VGPTQPGDVPY +R RCYE +++++ D
Sbjct 222 HRSHYELTKYALTKVQTESSKQPHLLLTPAVGPTQPGDVPYPVRVRCYEKILKYYGDD-- 279
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
ML LIP+ MRMAGPREC
Sbjct 280 -----------------------------------------EVMLALIPIPMRMAGPREC 298
Query 121 VLHARIRKNYGCTHFIVGRDHAGPSARRQDG 151
V HA IRKN+GCTHFIVGRDHAGPS ++G
Sbjct 299 VWHALIRKNFGCTHFIVGRDHAGPSTLTKNG 329
> sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step
of intracellular sulfate activation, essential for assimilatory
reduction of sulfate to sulfide, involved in methionine
metabolism (EC:2.7.7.4); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=511
Score = 92.8 bits (229), Expect = 4e-19, Method: Composition-based stats.
Identities = 57/144 (39%), Positives = 71/144 (49%), Gaps = 49/144 (34%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
HR+H EL RA + A A +L+ PVVG T+PGD+ ++ R R Y+ +I+
Sbjct 201 HRAHRELTVRAAREANAK------VLIHPVVGLTKPGDIDHHTRVRVYQEIIK------- 247
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
R P G A L L+PLAMRM+G RE
Sbjct 248 ---------------RYPNGI---------------------AFLSLLPLAMRMSGDREA 271
Query 121 VLHARIRKNYGCTHFIVGRDHAGP 144
V HA IRKNYG +HFIVGRDHAGP
Sbjct 272 VWHAIIRKNYGASHFIVGRDHAGP 295
> cel:T14G10.1 pps-1; 3'-Phosphoadenosine 5'-Phosphosulfate Synthetase
family member (pps-1); K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=652
Score = 48.5 bits (114), Expect = 9e-06, Method: Composition-based stats.
Identities = 42/143 (29%), Positives = 53/143 (37%), Gaps = 42/143 (29%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ R + A K P+LLL P+ G T+ DVP +R + +E VI DP
Sbjct 453 HNGH-ALLMRDTREKLLAEHKNPILLLHPLGGWTKDDDVPLDIRIKQHEAVIAERVLDPE 511
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
+L + P M AGP E
Sbjct 512 W-----------------------------------------TVLSIFPSPMMYAGPTEV 530
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
HAR R G H+IVGRD AG
Sbjct 531 QWHARSRIAAGIQHYIVGRDPAG 553
> ath:AT5G43780 APS4; APS4; sulfate adenylyltransferase (ATP);
K13811 3'-phosphoadenosine 5'-phosphosulfate synthase [EC:2.7.7.4
2.7.1.25]
Length=469
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 40/151 (26%), Positives = 54/151 (35%), Gaps = 44/151 (29%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ + K P+LLL P+ G T+ DVP R R +E V+ DP
Sbjct 257 HNGHALLMTDTRRRLLEMGYKNPVLLLNPLGGFTKADDVPLSWRMRQHEKVLEDGVLDPE 316
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 317 -----------------------------------------TTVVSIFPSPMLYAGPTEV 335
Query 121 VLHARIRKNYGCTHFIVGRDHAG---PSARR 148
HA+ R N G +IVGRD AG P+ +R
Sbjct 336 QWHAKARINAGANFYIVGRDPAGMGHPTEKR 366
> ath:AT4G14680 APS3; APS3; sulfate adenylyltransferase (ATP);
K13811 3'-phosphoadenosine 5'-phosphosulfate synthase [EC:2.7.7.4
2.7.1.25]
Length=465
Score = 45.1 bits (105), Expect = 9e-05, Method: Composition-based stats.
Identities = 37/143 (25%), Positives = 50/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ + K P+LLL P+ G T+ DVP R + +E V+ DP
Sbjct 255 HNGHALLMTDTRRRLLEMGYKNPILLLHPLGGFTKADDVPLSWRMKQHEKVLEDGVLDPE 314
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 315 -----------------------------------------TTVVSIFPSPMLYAGPTEV 333
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
HA+ R N G +IVGRD AG
Sbjct 334 QWHAKARINAGANFYIVGRDPAG 356
> dre:777719 papss2a, zgc:153748; 3'-phosphoadenosine 5'-phosphosulfate
synthase 2a; K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=612
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/143 (25%), Positives = 51/143 (35%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ + +RP+LLL P+ G T+ DVP R + + V+ DP
Sbjct 412 HNGHALLMTDTRRRINERGYRRPVLLLHPLGGWTKDDDVPLEWRMKQHAAVMEDGVLDPK 471
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++++ + P M AGP E
Sbjct 472 -----------------------------------------SSIVAIFPSPMMYAGPTEV 490
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R GC +IVGRD AG
Sbjct 491 QWHCRARMVAGCNFYIVGRDPAG 513
> ath:AT1G19920 APS2; APS2; sulfate adenylyltransferase (ATP);
K13811 3'-phosphoadenosine 5'-phosphosulfate synthase [EC:2.7.7.4
2.7.1.25]
Length=476
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 38/151 (25%), Positives = 53/151 (35%), Gaps = 44/151 (29%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ + K P+LLL P+ G T+ DVP +R + V+ DP
Sbjct 267 HNGHALLMNDTRKRLLEMGYKNPVLLLHPLGGFTKADDVPLDVRMEQHSKVLEDGVLDPK 326
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 327 -----------------------------------------TTIVSIFPSPMHYAGPTEV 345
Query 121 VLHARIRKNYGCTHFIVGRDHAG---PSARR 148
HA+ R N G +IVGRD AG P+ +R
Sbjct 346 QWHAKARINAGANFYIVGRDPAGMGHPTEKR 376
> dre:80939 papss2b, Papss1, id:ibd2761, papss2, sb:cb868, wu:fb12e05,
zgc:55851, zgc:85655; 3'-phosphoadenosine 5'-phosphosulfate
synthase 2b; K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=614
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 37/143 (25%), Positives = 50/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + K+P+LLL P+ G T+ DVP R R + V+ DP
Sbjct 414 HNGHALLMQDTKRRLLDRGYKKPVLLLHPLGGWTKEDDVPLDWRMRQHAAVLEEGVLDPE 473
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
N ++ + P M AGP E
Sbjct 474 -----------------------------------------NTIVAIFPSPMMYAGPTEV 492
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 493 QWHCRARMIAGANFYIVGRDPAG 515
> ath:AT3G22890 APS1; APS1 (ATP SULFURYLASE 1); sulfate adenylyltransferase
(ATP); K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=463
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 37/143 (25%), Positives = 50/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L+ + K P+LLL P+ G T+ DVP R + +E V+ DP
Sbjct 253 HNGHALLMTDTRRRLLEMGYKNPILLLHPLGGFTKADDVPLDWRMKQHEKVLEDGVLDPE 312
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 313 -----------------------------------------TTVVSIFPSPMHYAGPTEV 331
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
HA+ R N G +IVGRD AG
Sbjct 332 QWHAKARINAGANFYIVGRDPAG 354
> xla:398910 papss2, MGC68677; 3'-phosphoadenosine 5'-phosphosulfate
synthase 2; K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=621
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 36/143 (25%), Positives = 52/143 (36%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ +H + K P+LLL P+ G T+ DVP R + ++ V++ DP
Sbjct 421 HNGHALLMQDTRRHLLSRGYKCPVLLLHPLGGWTKDDDVPLDWRMKQHDAVLKEGVLDP- 479
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 480 ----------------------------------------KTTIVAIFPSPMLYAGPTEV 499
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 500 QWHCRARMIAGSNFYIVGRDPAG 522
> xla:735160 papss1, MGC114937, atpsk1, papss, sk1; 3'-phosphoadenosine
5'-phosphosulfate synthase 1 (EC:2.7.7.4); K13811
3'-phosphoadenosine 5'-phosphosulfate synthase [EC:2.7.7.4 2.7.1.25]
Length=624
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 35/143 (24%), Positives = 50/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + +RP+LLL P+ G T+ DVP R + + V++ DP
Sbjct 425 HNGHALLMQDTHKQLLERGYRRPVLLLHPLGGWTKDDDVPLMWRMKQHAAVLKEGVLDPE 484
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 485 -----------------------------------------TTVVAIFPSPMMYAGPTEV 503
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 504 QWHCRARMVAGANFYIVGRDPAG 526
> dre:559488 papss1, wu:fc43e12, zgc:194984, zgc:194985; 3'-phosphoadenosine
5'-phosphosulfate synthase 1 (EC:2.7.7.4); K13811
3'-phosphoadenosine 5'-phosphosulfate synthase [EC:2.7.7.4
2.7.1.25]
Length=624
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 35/143 (24%), Positives = 51/143 (35%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + +RP+LLL P+ G T+ DVP R + + V+ DP+
Sbjct 425 HNGHALLMQDTQRRLIERGYRRPVLLLHPLGGWTKDDDVPLAWRMKQHAAVLEEGLLDPN 484
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
+ ++ + P M AGP E
Sbjct 485 -----------------------------------------STIVAIFPSPMMYAGPTEV 503
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 504 QWHCRARMVAGANFYIVGRDPAG 526
> mmu:23971 Papss1, AI325286, Asapk, SK1; 3'-phosphoadenosine
5'-phosphosulfate synthase 1 (EC:2.7.1.25 2.7.7.4); K13811 3'-phosphoadenosine
5'-phosphosulfate synthase [EC:2.7.7.4 2.7.1.25]
Length=624
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 35/143 (24%), Positives = 49/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + +RP+LLL P+ G T+ DVP R + + V+ DP
Sbjct 425 HNGHALLMQDTHKQLLERGYRRPVLLLHPLGGWTKDDDVPLMWRMKQHAAVLEEGILDPE 484
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 485 -----------------------------------------TTVVAIFPSPMMYAGPTEV 503
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 504 QWHCRARMVAGANFYIVGRDPAG 526
> hsa:9061 PAPSS1, ATPSK1, PAPSS, SK1; 3'-phosphoadenosine 5'-phosphosulfate
synthase 1 (EC:2.7.1.25 2.7.7.4); K13811 3'-phosphoadenosine
5'-phosphosulfate synthase [EC:2.7.7.4 2.7.1.25]
Length=624
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 34/143 (23%), Positives = 49/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + +RP+LLL P+ G T+ DVP R + + V+ +P
Sbjct 425 HNGHALLMQDTHKQLLERGYRRPVLLLHPLGGWTKDDDVPLMWRMKQHAAVLEEGVLNPE 484
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
++ + P M AGP E
Sbjct 485 -----------------------------------------TTVVAIFPSPMMYAGPTEV 503
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 504 QWHCRARMVAGANFYIVGRDPAG 526
> mmu:23972 Papss2, 1810018P12Rik, AI159688, AtpsU2, Atpsk2, Sk2,
bm; 3'-phosphoadenosine 5'-phosphosulfate synthase 2 (EC:2.7.1.25
2.7.7.4); K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=614
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/143 (24%), Positives = 49/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + K P+LLL P+ G T+ DVP R + + V+ DP
Sbjct 414 HNGHALLMQDTRRRLLERGYKHPVLLLHPLGGWTKDDDVPLEWRMKQHAAVLEERVLDPK 473
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
+ ++ + P M AGP E
Sbjct 474 -----------------------------------------STIVAIFPSPMLYAGPTEV 492
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 493 QWHCRCRMIAGANFYIVGRDPAG 515
> hsa:9060 PAPSS2, ATPSK2, SK2; 3'-phosphoadenosine 5'-phosphosulfate
synthase 2 (EC:2.7.1.25 2.7.7.4); K13811 3'-phosphoadenosine
5'-phosphosulfate synthase [EC:2.7.7.4 2.7.1.25]
Length=619
Score = 39.3 bits (90), Expect = 0.005, Method: Composition-based stats.
Identities = 35/143 (24%), Positives = 49/143 (34%), Gaps = 41/143 (28%)
Query 1 HRSHYELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVPYYLRARCYEHVIRHFEQDPS 60
H H L++ + K P+LLL P+ G T+ DVP R + + V+ DP
Sbjct 420 HNGHALLMQDTRRRLLERGYKHPVLLLHPLGGWTKDDDVPLDWRMKQHAAVLEEGVLDPK 479
Query 61 CQFPHLCPTSASTPARSPRGHQQQQQQQQQQEQQGPSSSSSNAMLVLIPLAMRMAGPREC 120
+ ++ + P M AGP E
Sbjct 480 -----------------------------------------STIVAIFPSPMLYAGPTEV 498
Query 121 VLHARIRKNYGCTHFIVGRDHAG 143
H R R G +IVGRD AG
Sbjct 499 QWHCRSRMIAGANFYIVGRDPAG 521
> dre:566433 tight junction protein 1-like; K05701 tight junction
protein 1
Length=1544
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 2/76 (2%)
Query 6 ELVKRALQHAAAAAKKRPMLLLTPVVGPTQPGDVP-YYLRARCYEHVIRHFEQDPSCQFP 64
E ++ L + + +P LTP + P QP P +Y RA E + +P+ P
Sbjct 1204 EYYRKQLSYFDRRSFDKPSTQLTPAIKPAQPQTQPAHYPRAESVEKINPAAPSNPAVPPP 1263
Query 65 HLCPTSASTPARSPRG 80
L P A+TP P G
Sbjct 1264 TL-PKPAATPPLDPHG 1278
Lambda K H
0.321 0.132 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40