bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7785_orf1
Length=89
Score E
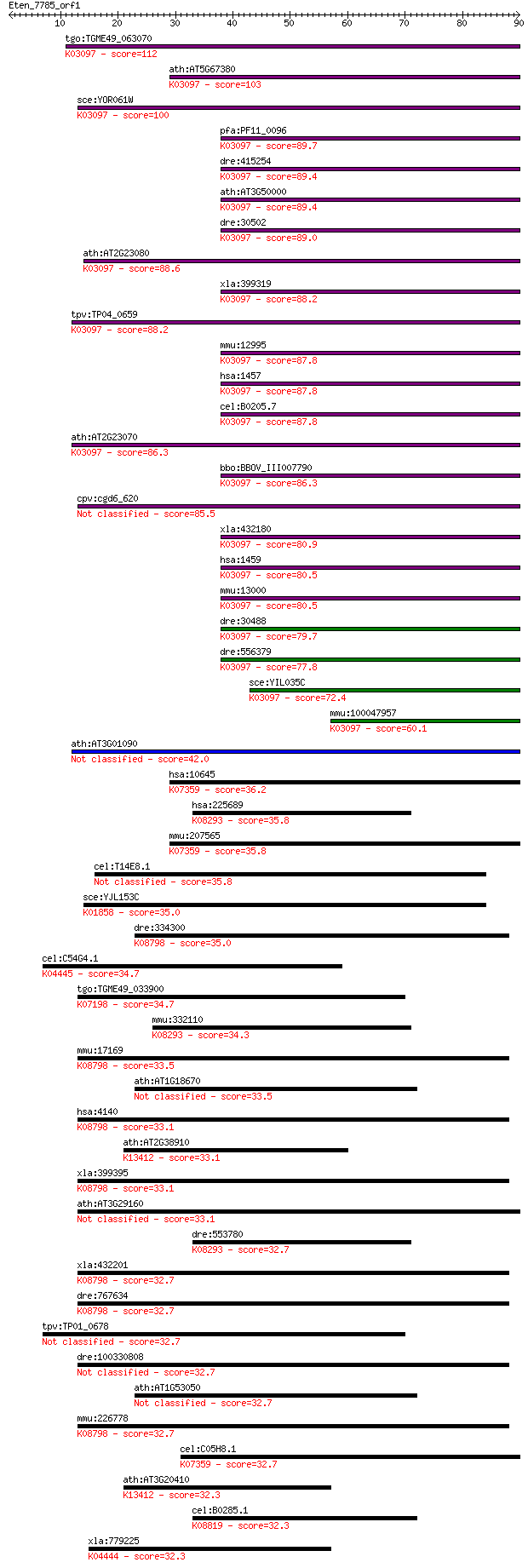
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_063070 casein kinase II alpha subunit, putative (EC... 112 3e-25
ath:AT5G67380 CKA1; CKA1 (CASEIN KINASE ALPHA 1); kinase; K030... 103 2e-22
sce:YOR061W CKA2, YOR29-12; Alpha' catalytic subunit of casein... 100 9e-22
pfa:PF11_0096 CK2alpha; casein kinase II, alpha subunit; K0309... 89.7 2e-18
dre:415254 ck2a1, im:6891713; zgc:86598 (EC:2.7.11.1); K03097 ... 89.4 2e-18
ath:AT3G50000 CKA2; CKA2 (CASEIN KINASE II, ALPHA CHAIN 2); ki... 89.4 3e-18
dre:30502 ck2a1, wu:fi38e04, wu:fi38h03; casein kinase 2 alpha... 89.0 4e-18
ath:AT2G23080 casein kinase II alpha chain, putative; K03097 c... 88.6 4e-18
xla:399319 csnk2a1, ck2a1; casein kinase 2, alpha 1 polypeptid... 88.2 6e-18
tpv:TP04_0659 casein kinase II subunit alpha (EC:2.7.1.37); K0... 88.2 6e-18
mmu:12995 Csnk2a1, Csnk2a1-rs4, MGC102141; casein kinase 2, al... 87.8 7e-18
hsa:1457 CSNK2A1, CK2A1, CKII; casein kinase 2, alpha 1 polype... 87.8 8e-18
cel:B0205.7 kin-3; protein KINase family member (kin-3); K0309... 87.8 8e-18
ath:AT2G23070 casein kinase II alpha chain, putative; K03097 c... 86.3 2e-17
bbo:BBOV_III007790 17.m07683; casein kinase II, alpha chain (C... 86.3 2e-17
cpv:cgd6_620 casein kinase II, alpha subunit 85.5 4e-17
xla:432180 csnk2a2, MGC83125; casein kinase 2, alpha prime pol... 80.9 9e-16
hsa:1459 CSNK2A2, CK2A2, CSNK2A1, FLJ43934; casein kinase 2, a... 80.5 1e-15
mmu:13000 Csnk2a2, 1110035J23Rik, C77789, CK2; casein kinase 2... 80.5 1e-15
dre:30488 ck2a2a, ck2a2, wu:fi09h12; casein kinase 2 alpha 2a;... 79.7 2e-15
dre:556379 ck2a2b; casein kinase 2 alpha 2b; K03097 casein kin... 77.8 7e-15
sce:YIL035C CKA1; Alpha catalytic subunit of casein kinase 2, ... 72.4 3e-13
mmu:100047957 casein kinase II subunit alpha-like; K03097 case... 60.1 2e-09
ath:AT3G01090 AKIN10; AKIN10 (Arabidopsis SNF1 kinase homolog ... 42.0 5e-04
hsa:10645 CAMKK2, CAMKK, CAMKKB, KIAA0787, MGC15254; calcium/c... 36.2 0.027
hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinas... 35.8 0.031
mmu:207565 Camkk2, 6330570N16Rik, AW061083, mKIAA0787; calcium... 35.8 0.033
cel:T14E8.1 hypothetical protein 35.8 0.034
sce:YJL153C INO1, APR1; Ino1p (EC:5.5.1.4); K01858 myo-inosito... 35.0 0.061
dre:334300 mark3, MGC152848, fi39g08, wu:fi39g08, zgc:152848, ... 35.0 0.062
cel:C54G4.1 rskn-2; RSK-pNinety (RSK-p90 kinase) homolog famil... 34.7 0.077
tgo:TGME49_033900 serine/threonine-protein kinase, putative (E... 34.7 0.083
mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activ... 34.3 0.11
mmu:17169 Mark3, 1600015G02Rik, A430080F22Rik, C-TAK1, ETK-1, ... 33.5 0.17
ath:AT1G18670 IBS1; IBS1 (IMPAIRED IN BABA-INDUCED STERILITY 1... 33.5 0.19
hsa:4140 MARK3, CTAK1, KP78, PAR1A; MAP/microtubule affinity-r... 33.1 0.19
ath:AT2G38910 CPK20; CPK20; ATP binding / calcium ion binding ... 33.1 0.19
xla:399395 mark3, Par-1A, par-1, par1; MAP/microtubule affinit... 33.1 0.22
ath:AT3G29160 AKIN11; AKIN11 (Arabidopsis SNF1 kinase homolog ... 33.1 0.24
dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activa... 32.7 0.25
xla:432201 mark1, MGC80341; MAP/microtubule affinity-regulatin... 32.7 0.25
dre:767634 MGC153725; zgc:153725 (EC:2.7.11.1); K08798 MAP/mic... 32.7 0.26
tpv:TP01_0678 serine/threonine protein kinase 32.7 0.27
dre:100330808 MAP/microtubule affinity-regulating kinase 3-like 32.7 0.27
ath:AT1G53050 protein kinase family protein 32.7 0.28
mmu:226778 Mark1, AW491150, B930025N23Rik, Emk3, KIAA1477, mKI... 32.7 0.29
cel:C05H8.1 ckk-1; CaM Kinase Kinase family member (ckk-1); K0... 32.7 0.30
ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);... 32.3 0.34
cel:B0285.1 hypothetical protein; K08819 Cdc2-related kinase, ... 32.3 0.35
xla:779225 mapkapk3, MGC154360; mitogen-activated protein kina... 32.3 0.36
> tgo:TGME49_063070 casein kinase II alpha subunit, putative (EC:2.7.11.1
2.7.11.23); K03097 casein kinase II subunit alpha
[EC:2.7.11.1]
Length=539
Score = 112 bits (280), Expect = 3e-25, Method: Composition-based stats.
Identities = 62/79 (78%), Positives = 72/79 (91%), Gaps = 0/79 (0%)
Query 11 LAVVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVN 70
L G++CV+K+LKPVKKKKIRRE+KILQNL GGPNII+LLDVVKDP SRTPAL+FE++N
Sbjct 252 LGFEGEKCVVKILKPVKKKKIRREVKILQNLCGGPNIIRLLDVVKDPASRTPALVFEHIN 311
Query 71 NVDFKTLYPTLQDYEIRYY 89
N DFKTLYPTL DY+IRYY
Sbjct 312 NQDFKTLYPTLTDYDIRYY 330
> ath:AT5G67380 CKA1; CKA1 (CASEIN KINASE ALPHA 1); kinase; K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=376
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 48/61 (78%), Positives = 55/61 (90%), Gaps = 0/61 (0%)
Query 29 KKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRY 88
+KIRREIKILQNL GGPNI+KLLDVV+D S+TP+LIFEYVN+ DFK LYPTL DY+IRY
Sbjct 114 RKIRREIKILQNLCGGPNIVKLLDVVRDQHSKTPSLIFEYVNSTDFKVLYPTLTDYDIRY 173
Query 89 Y 89
Y
Sbjct 174 Y 174
> sce:YOR061W CKA2, YOR29-12; Alpha' catalytic subunit of casein
kinase 2, a Ser/Thr protein kinase with roles in cell growth
and proliferation; the holoenzyme also contains CKA1, CKB1
and CKB2, the many substrates include transcription factors
and all RNA polymerases (EC:2.7.11.1); K03097 casein kinase
II subunit alpha [EC:2.7.11.1]
Length=339
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 49/77 (63%), Positives = 61/77 (79%), Gaps = 0/77 (0%)
Query 13 VVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNV 72
V Q+CVIKVLKPVK KKI RE+KIL NL GGPN++ L D+V+D S+ PALIFE + NV
Sbjct 71 VNNQKCVIKVLKPVKMKKIYRELKILTNLTGGPNVVGLYDIVQDADSKIPALIFEEIKNV 130
Query 73 DFKTLYPTLQDYEIRYY 89
DF+TLYPT + +I+YY
Sbjct 131 DFRTLYPTFKLPDIQYY 147
> pfa:PF11_0096 CK2alpha; casein kinase II, alpha subunit; K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=335
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 39/52 (75%), Positives = 47/52 (90%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
LQNL GGPNIIKLLD+VKDP+++TP+LIFEY+NN+DFKTLYP D +IRYY
Sbjct 89 LQNLNGGPNIIKLLDIVKDPVTKTPSLIFEYINNIDFKTLYPKFTDKDIRYY 140
> dre:415254 ck2a1, im:6891713; zgc:86598 (EC:2.7.11.1); K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=393
Score = 89.4 bits (220), Expect = 2e-18, Method: Composition-based stats.
Identities = 38/52 (73%), Positives = 46/52 (88%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GGPNII LLD++KDP+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 85 LENLRGGPNIITLLDIIKDPVSRTPALVFEHVNNTDFKQLYQTLSDYDIRFY 136
> ath:AT3G50000 CKA2; CKA2 (CASEIN KINASE II, ALPHA CHAIN 2);
kinase; K03097 casein kinase II subunit alpha [EC:2.7.11.1]
Length=403
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 40/52 (76%), Positives = 46/52 (88%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
LQNL GGPNI+KLLDVV+D S+TP+LIFEYVN+ DFK LYPTL DY+IRYY
Sbjct 150 LQNLCGGPNIVKLLDVVRDQHSKTPSLIFEYVNSTDFKVLYPTLTDYDIRYY 201
> dre:30502 ck2a1, wu:fi38e04, wu:fi38h03; casein kinase 2 alpha
1; K03097 casein kinase II subunit alpha [EC:2.7.11.1]
Length=395
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 38/52 (73%), Positives = 46/52 (88%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GGPNII LLD++KDP+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 85 LENLRGGPNIITLLDIIKDPVSRTPALVFEHVNNTDFKQLYQTLSDYDIRFY 136
> ath:AT2G23080 casein kinase II alpha chain, putative; K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=333
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 56/76 (73%), Positives = 68/76 (89%), Gaps = 0/76 (0%)
Query 14 VGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVD 73
+RCVIK+LKPVKKKKI+REIKILQNL GGPNI+KL D+V+D S+TP+L+FE+VN+VD
Sbjct 56 TNERCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLYDIVRDEHSKTPSLVFEFVNSVD 115
Query 74 FKTLYPTLQDYEIRYY 89
FK LYPTL DY+IRYY
Sbjct 116 FKVLYPTLTDYDIRYY 131
> xla:399319 csnk2a1, ck2a1; casein kinase 2, alpha 1 polypeptide
(EC:2.7.11.1); K03097 casein kinase II subunit alpha [EC:2.7.11.1]
Length=401
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 38/52 (73%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GGPNII L D+VKDP+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 85 LENLRGGPNIITLADIVKDPVSRTPALVFEHVNNTDFKQLYQTLTDYDIRFY 136
> tpv:TP04_0659 casein kinase II subunit alpha (EC:2.7.1.37);
K03097 casein kinase II subunit alpha [EC:2.7.11.1]
Length=420
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 60/78 (76%), Positives = 68/78 (87%), Gaps = 0/78 (0%)
Query 12 AVVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN 71
V +CVIK+LKPVKKKKI+REIKILQNL GGPNIIKLLD+VKDP SRTP+LIFE+VNN
Sbjct 147 TVTKDKCVIKILKPVKKKKIKREIKILQNLRGGPNIIKLLDIVKDPQSRTPSLIFEHVNN 206
Query 72 VDFKTLYPTLQDYEIRYY 89
DFKTLYPTL +I+YY
Sbjct 207 TDFKTLYPTLSIQDIKYY 224
> mmu:12995 Csnk2a1, Csnk2a1-rs4, MGC102141; casein kinase 2,
alpha 1 polypeptide (EC:2.7.11.1); K03097 casein kinase II subunit
alpha [EC:2.7.11.1]
Length=391
Score = 87.8 bits (216), Expect = 7e-18, Method: Composition-based stats.
Identities = 38/52 (73%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GGPNII L D+VKDP+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 85 LENLRGGPNIITLADIVKDPVSRTPALVFEHVNNTDFKQLYQTLTDYDIRFY 136
> hsa:1457 CSNK2A1, CK2A1, CKII; casein kinase 2, alpha 1 polypeptide
(EC:2.7.11.1); K03097 casein kinase II subunit alpha
[EC:2.7.11.1]
Length=391
Score = 87.8 bits (216), Expect = 8e-18, Method: Composition-based stats.
Identities = 38/52 (73%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GGPNII L D+VKDP+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 85 LENLRGGPNIITLADIVKDPVSRTPALVFEHVNNTDFKQLYQTLTDYDIRFY 136
> cel:B0205.7 kin-3; protein KINase family member (kin-3); K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=360
Score = 87.8 bits (216), Expect = 8e-18, Method: Composition-based stats.
Identities = 41/52 (78%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NII LLDVVKDP+SRTPALIFE+VNN DFK LY TL DY+IRYY
Sbjct 84 LENLRGGTNIITLLDVVKDPISRTPALIFEHVNNSDFKQLYQTLSDYDIRYY 135
> ath:AT2G23070 casein kinase II alpha chain, putative; K03097
casein kinase II subunit alpha [EC:2.7.11.1]
Length=432
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 57/78 (73%), Positives = 69/78 (88%), Gaps = 0/78 (0%)
Query 12 AVVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN 71
A ++CVIK+LKPVKKKKI+REIKILQNL GGPNI+KLLD+V+D S+TP+LIFE+VNN
Sbjct 152 ATDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLLDIVRDQQSKTPSLIFEHVNN 211
Query 72 VDFKTLYPTLQDYEIRYY 89
DFK LYPTL DY++RYY
Sbjct 212 KDFKVLYPTLSDYDVRYY 229
> bbo:BBOV_III007790 17.m07683; casein kinase II, alpha chain
(CK II) (EC:2.7.11.1); K03097 casein kinase II subunit alpha
[EC:2.7.11.1]
Length=423
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 40/52 (76%), Positives = 44/52 (84%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
LQNL GGPNIIKLLD+VKDP SRTP+LIFEYVNN DFK LYPT +I+YY
Sbjct 180 LQNLSGGPNIIKLLDIVKDPQSRTPSLIFEYVNNTDFKILYPTFTIADIKYY 231
> cpv:cgd6_620 casein kinase II, alpha subunit
Length=341
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/77 (61%), Positives = 65/77 (84%), Gaps = 0/77 (0%)
Query 13 VVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNV 72
+ ++ +IKVL+PVK+KKI+REIKIL NL GGPNI+KLLD++KDP S T +L+FE ++NV
Sbjct 65 ITNEKVIIKVLRPVKRKKIKREIKILYNLHGGPNIVKLLDIIKDPSSGTISLVFECLDNV 124
Query 73 DFKTLYPTLQDYEIRYY 89
DFK+L+PT D +IR+Y
Sbjct 125 DFKSLFPTFNDNDIRFY 141
> xla:432180 csnk2a2, MGC83125; casein kinase 2, alpha prime polypeptide
(EC:2.7.11.1); K03097 casein kinase II subunit alpha
[EC:2.7.11.1]
Length=349
Score = 80.9 bits (198), Expect = 9e-16, Method: Composition-based stats.
Identities = 35/52 (67%), Positives = 44/52 (84%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NII+LLD +KDP+S+TPAL+FEY+NN DFK LY L DY+IR+Y
Sbjct 86 LENLRGGTNIIRLLDTIKDPVSKTPALVFEYINNTDFKQLYQILTDYDIRFY 137
> hsa:1459 CSNK2A2, CK2A2, CSNK2A1, FLJ43934; casein kinase 2,
alpha prime polypeptide (EC:2.7.11.1); K03097 casein kinase
II subunit alpha [EC:2.7.11.1]
Length=350
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 35/52 (67%), Positives = 44/52 (84%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NIIKL+D VKDP+S+TPAL+FEY+NN DFK LY L D++IR+Y
Sbjct 86 LENLRGGTNIIKLIDTVKDPVSKTPALVFEYINNTDFKQLYQILTDFDIRFY 137
> mmu:13000 Csnk2a2, 1110035J23Rik, C77789, CK2; casein kinase
2, alpha prime polypeptide (EC:2.7.11.1); K03097 casein kinase
II subunit alpha [EC:2.7.11.1]
Length=350
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 35/52 (67%), Positives = 44/52 (84%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NIIKL+D VKDP+S+TPAL+FEY+NN DFK LY L D++IR+Y
Sbjct 86 LENLRGGTNIIKLIDTVKDPVSKTPALVFEYINNTDFKQLYQILTDFDIRFY 137
> dre:30488 ck2a2a, ck2a2, wu:fi09h12; casein kinase 2 alpha 2a;
K03097 casein kinase II subunit alpha [EC:2.7.11.1]
Length=348
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 35/52 (67%), Positives = 44/52 (84%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NII+L+D VKDP+SRTPAL+FEY+NN DFK LY L D++IR+Y
Sbjct 85 LENLRGGTNIIRLVDTVKDPVSRTPALVFEYINNTDFKDLYQRLTDFDIRFY 136
> dre:556379 ck2a2b; casein kinase 2 alpha 2b; K03097 casein kinase
II subunit alpha [EC:2.7.11.1]
Length=350
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 35/52 (67%), Positives = 43/52 (82%), Gaps = 0/52 (0%)
Query 38 LQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
L+NL GG NII+L+D VKDP+SRTPAL+FE +NN DFK LY L DY+IR+Y
Sbjct 86 LENLRGGTNIIQLMDTVKDPVSRTPALVFECINNTDFKELYQKLTDYDIRFY 137
> sce:YIL035C CKA1; Alpha catalytic subunit of casein kinase 2,
a Ser/Thr protein kinase with roles in cell growth and proliferation;
the holoenzyme also contains CKA2, CKB1 and CKB2,
the many substrates include transcription factors and all
RNA polymerases (EC:2.7.11.1); K03097 casein kinase II subunit
alpha [EC:2.7.11.1]
Length=372
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 31/47 (65%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 43 GGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
G NII L D++KDP+S+TPAL+FEYV+NVDF+ LYP L D EIR+Y
Sbjct 129 GHANIIHLFDIIKDPISKTPALVFEYVDNVDFRILYPKLTDLEIRFY 175
> mmu:100047957 casein kinase II subunit alpha-like; K03097 casein
kinase II subunit alpha [EC:2.7.11.1]
Length=331
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 29/33 (87%), Gaps = 0/33 (0%)
Query 57 PLSRTPALIFEYVNNVDFKTLYPTLQDYEIRYY 89
P+SRTPAL+FE+VNN DFK LY TL DY+IR+Y
Sbjct 44 PVSRTPALVFEHVNNTDFKQLYQTLTDYDIRFY 76
> ath:AT3G01090 AKIN10; AKIN10 (Arabidopsis SNF1 kinase homolog
10); protein binding / protein kinase
Length=512
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 33/87 (37%), Positives = 48/87 (55%), Gaps = 12/87 (13%)
Query 12 AVVGQRCVIKVLKPVKKK------KIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALI 65
A+ G + IK+L K K K+RREIKIL+ LF P+II+L +V++ P L+
Sbjct 39 ALTGHKVAIKILNRRKIKNMEMEEKVRREIKILR-LFMHPHIIRLYEVIETPTD--IYLV 95
Query 66 FEYVNN---VDFKTLYPTLQDYEIRYY 89
EYVN+ D+ LQ+ E R +
Sbjct 96 MEYVNSGELFDYIVEKGRLQEDEARNF 122
> hsa:10645 CAMKK2, CAMKK, CAMKKB, KIAA0787, MGC15254; calcium/calmodulin-dependent
protein kinase kinase 2, beta (EC:2.7.11.17);
K07359 calcium/calmodulin-dependent protein kinase kinase
[EC:2.7.11.17]
Length=588
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 29 KKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN---VDFKTLYPTLQDYE 85
+++ +EI IL+ L PN++KL++V+ DP ++FE VN ++ TL P +D +
Sbjct 231 EQVYQEIAILKKL-DHPNVVKLVEVLDDPNEDHLYMVFELVNQGPVMEVPTLKPLSED-Q 288
Query 86 IRYY 89
R+Y
Sbjct 289 ARFY 292
> hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinase
15 (EC:2.7.11.24); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=544
Score = 35.8 bits (81), Expect = 0.031, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 33 REIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVN 70
REI +LQ PNII LLDV++ R L+FE+++
Sbjct 59 REITLLQEFGDHPNIISLLDVIRAENDRDIYLVFEFMD 96
> mmu:207565 Camkk2, 6330570N16Rik, AW061083, mKIAA0787; calcium/calmodulin-dependent
protein kinase kinase 2, beta (EC:2.7.11.17);
K07359 calcium/calmodulin-dependent protein kinase
kinase [EC:2.7.11.17]
Length=588
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 29 KKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN---VDFKTLYPTLQDYE 85
+++ +EI IL+ L PN++KL++V+ DP ++FE VN ++ TL P +D +
Sbjct 231 EQVYQEIAILKKL-DHPNVVKLVEVLDDPNEDHLYMVFELVNQGPVMEVPTLKPLSED-Q 288
Query 86 IRYY 89
R+Y
Sbjct 289 ARFY 292
> cel:T14E8.1 hypothetical protein
Length=1086
Score = 35.8 bits (81), Expect = 0.034, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 1/68 (1%)
Query 16 QRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNNVDFK 75
++ V K LK K + E + + F PNI+KL+ V D S P +I EY+ D K
Sbjct 762 EKVVCKYLKEGKISEFYEEARTMSE-FDHPNILKLIGVALDDSSHLPIIITEYMAKGDLK 820
Query 76 TLYPTLQD 83
+ +++
Sbjct 821 SFIENVEN 828
> sce:YJL153C INO1, APR1; Ino1p (EC:5.5.1.4); K01858 myo-inositol-1-phosphate
synthase [EC:5.5.1.4]
Length=533
Score = 35.0 bits (79), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 5/75 (6%)
Query 14 VGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALI-----FEY 68
V VIK +KPV K+ + + + GG N I + +V +D L TP +I E+
Sbjct 396 VDHCIVIKYMKPVGDSKVAMDEYYSELMLGGHNRISIHNVCEDSLLATPLIIDLLVMTEF 455
Query 69 VNNVDFKTLYPTLQD 83
V +K + P +D
Sbjct 456 CTRVSYKKVDPVKED 470
> dre:334300 mark3, MGC152848, fi39g08, wu:fi39g08, zgc:152848,
zgc:55693; MAP/microtubule affinity-regulating kinase 3 (EC:2.7.11.1);
K08798 MAP/microtubule affinity-regulating kinase
[EC:2.7.11.1]
Length=722
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 38/68 (55%), Gaps = 6/68 (8%)
Query 23 LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN---VDFKTLYP 79
L P +K+ RE+ I++NL PNI+KL +V++ +T L+ EY + D+ +
Sbjct 93 LNPTSLQKLSREVTIMKNL-NHPNIVKLFEVIET--EKTLFLVMEYASGGEVFDYLVAHG 149
Query 80 TLQDYEIR 87
+++ E R
Sbjct 150 RMKEKEAR 157
> cel:C54G4.1 rskn-2; RSK-pNinety (RSK-p90 kinase) homolog family
member (rskn-2); K04445 mitogen-,stress activated protein
kinases [EC:2.7.11.1]
Length=772
Score = 34.7 bits (78), Expect = 0.077, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 7 QRKSLAVVGQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPL 58
+R V G + +K++ + +RE +IL+ + G PNI++L DV DPL
Sbjct 397 RRCERVVDGAQFAVKIVSQKFASQAQREARILEMVQGHPNIVQLHDVHSDPL 448
> tgo:TGME49_033900 serine/threonine-protein kinase, putative
(EC:2.7.11.17); K07198 5'-AMP-activated protein kinase, catalytic
alpha subunit [EC:2.7.11.11]
Length=412
Score = 34.7 bits (78), Expect = 0.083, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 9/63 (14%)
Query 13 VVGQRCVIKVLKPVKKK------KIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIF 66
V GQ+ +K++ K + KIRREI ILQ L P++I+L +++ P ++
Sbjct 86 VTGQKVAVKIINKAKMEMMEMYEKIRREINILQCLH-HPHVIRLYELIDTPTD--IFMVM 142
Query 67 EYV 69
EYV
Sbjct 143 EYV 145
> mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=549
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 26 VKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVN 70
+ ++ REI +L+ G PNII+LLDV+ R L+FE ++
Sbjct 53 IDAQRTFREIMLLKEFGGHPNIIRLLDVIPAKNDRDIYLVFESMD 97
> mmu:17169 Mark3, 1600015G02Rik, A430080F22Rik, C-TAK1, ETK-1,
ETK1, Emk2, KIAA4230, MPK10; MAP/microtubule affinity-regulating
kinase 3 (EC:2.7.11.1); K08798 MAP/microtubule affinity-regulating
kinase [EC:2.7.11.1]
Length=744
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ IK+ L P +K+ RE++I++ + PNI+KL +V++ +T LI E
Sbjct 77 LTGREVAIKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--EKTLYLIME 133
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 134 YASGGEVFDYLVAHGRMKEKEAR 156
> ath:AT1G18670 IBS1; IBS1 (IMPAIRED IN BABA-INDUCED STERILITY
1); ATP binding / kinase/ protein kinase/ protein serine/threonine
kinase/ protein tyrosine kinase
Length=709
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 23 LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN 71
+P + + REI IL+ L PNIIKL +V LS + L+FEY+ +
Sbjct 167 FEPESVRFMAREILILRKL-NHPNIIKLEGIVTSKLSCSIHLVFEYMEH 214
> hsa:4140 MARK3, CTAK1, KP78, PAR1A; MAP/microtubule affinity-regulating
kinase 3 (EC:2.7.11.1); K08798 MAP/microtubule affinity-regulating
kinase [EC:2.7.11.1]
Length=753
Score = 33.1 bits (74), Expect = 0.19, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ IK+ L P +K+ RE++I++ + PNI+KL +V++ +T LI E
Sbjct 77 LTGREVAIKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--EKTLYLIME 133
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 134 YASGGEVFDYLVAHGRMKEKEAR 156
> ath:AT2G38910 CPK20; CPK20; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=583
Score = 33.1 bits (74), Expect = 0.19, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 21 KVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLS 59
K+ P + +RREI+I+ +L G PN+I+++ +D ++
Sbjct 169 KLTTPEDVEDVRREIQIMHHLSGHPNVIQIVGAYEDAVA 207
> xla:399395 mark3, Par-1A, par-1, par1; MAP/microtubule affinity-regulating
kinase 3 (EC:2.7.11.1); K08798 MAP/microtubule
affinity-regulating kinase [EC:2.7.11.1]
Length=725
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ IK+ L P +K+ RE++I++ + PNI+KL +V++ +T LI E
Sbjct 77 LTGREVAIKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--EKTLYLIME 133
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 134 YASGGEVFDYLVAHGRMKEKEAR 156
> ath:AT3G29160 AKIN11; AKIN11 (Arabidopsis SNF1 kinase homolog
11); protein binding / protein kinase
Length=512
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 45/86 (52%), Gaps = 12/86 (13%)
Query 13 VVGQRCVIKVLKPVKKK------KIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIF 66
V G + IK+L K K K+RREIKIL+ LF P+II+ +V++ + ++
Sbjct 41 VTGHKVAIKILNRRKIKNMEMEEKVRREIKILR-LFMHPHIIRQYEVIET--TSDIYVVM 97
Query 67 EYVNN---VDFKTLYPTLQDYEIRYY 89
EYV + D+ LQ+ E R +
Sbjct 98 EYVKSGELFDYIVEKGRLQEDEARNF 123
> dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=524
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 33 REIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVN 70
REI LQ PNIIKLL+V++ + LIFE+++
Sbjct 60 REIMFLQEFGDHPNIIKLLNVIRAQNDKDIYLIFEFMD 97
> xla:432201 mark1, MGC80341; MAP/microtubule affinity-regulating
kinase 1 (EC:2.7.11.1); K08798 MAP/microtubule affinity-regulating
kinase [EC:2.7.11.1]
Length=792
Score = 32.7 bits (73), Expect = 0.25, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ +K+ L P +K+ RE++I++ + PNI+KL +V++ +T LI E
Sbjct 81 LTGREVAVKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--EKTLYLIME 137
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 138 YASGGEVFDYLVAHGRMKEKEAR 160
> dre:767634 MGC153725; zgc:153725 (EC:2.7.11.1); K08798 MAP/microtubule
affinity-regulating kinase [EC:2.7.11.1]
Length=754
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ IK+ L P +K+ RE++I++ + PNI+KL +V++ +T L+ E
Sbjct 77 LTGREVAIKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--DKTLYLVME 133
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 134 YASGGEVFDYLVAHGRMKEKEAR 156
> tpv:TP01_0678 serine/threonine protein kinase
Length=677
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 32/64 (50%), Gaps = 1/64 (1%)
Query 7 QRKSLAVVGQRCVIKVLK-PVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALI 65
+R + VV + + + ++ REI LQ L PNI+KL DV +R L+
Sbjct 34 KRDTNEVVALKKIFDAFRNSTDAQRTYREIMFLQKLKKCPNIVKLRDVYPADNNRDVYLV 93
Query 66 FEYV 69
FEYV
Sbjct 94 FEYV 97
> dre:100330808 MAP/microtubule affinity-regulating kinase 3-like
Length=527
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ IK+ L P +K+ RE++I++ + PNI+KL +V++ +T L+ E
Sbjct 79 LTGREVAIKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--DKTLYLVME 135
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 136 YASGGEVFDYLVAHGRMKEKEAR 158
> ath:AT1G53050 protein kinase family protein
Length=694
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 23 LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN 71
L+P + + REI+IL+ L PNIIKL +V +S + L+FEY+ +
Sbjct 170 LEPESVRFMAREIQILRRL-DHPNIIKLEGLVTSRMSCSLYLVFEYMEH 217
> mmu:226778 Mark1, AW491150, B930025N23Rik, Emk3, KIAA1477, mKIAA1477;
MAP/microtubule affinity-regulating kinase 1 (EC:2.7.11.1);
K08798 MAP/microtubule affinity-regulating kinase
[EC:2.7.11.1]
Length=795
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 44/83 (53%), Gaps = 11/83 (13%)
Query 13 VVGQRCVIKV-----LKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFE 67
+ G+ +K+ L P +K+ RE++I++ + PNI+KL +V++ +T L+ E
Sbjct 81 LTGREVAVKIIDKTQLNPTSLQKLFREVRIMK-ILNHPNIVKLFEVIET--EKTLYLVME 137
Query 68 YVNN---VDFKTLYPTLQDYEIR 87
Y + D+ + +++ E R
Sbjct 138 YASGGEVFDYLVAHGRMKEKEAR 160
> cel:C05H8.1 ckk-1; CaM Kinase Kinase family member (ckk-1);
K07359 calcium/calmodulin-dependent protein kinase kinase [EC:2.7.11.17]
Length=541
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 35/62 (56%), Gaps = 4/62 (6%)
Query 31 IRREIKILQNLFGGPNIIKLLDVVKDPLSRTPALIFEYVNN---VDFKTLYPTLQDYEIR 87
+++EI IL+ L PN++KL++V+ DP ++FE+V ++ T P +D
Sbjct 197 VQKEIAILKKL-SHPNVVKLVEVLDDPNDNYLYMVFEFVEKGSILEIPTDKPLDEDTAWS 255
Query 88 YY 89
Y+
Sbjct 256 YF 257
> ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=541
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 21 KVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKD 56
K++ K +RREI+I+Q+L G PNI++ +D
Sbjct 126 KLVTKADKDDMRREIQIMQHLSGQPNIVEFKGAYED 161
> cel:B0285.1 hypothetical protein; K08819 Cdc2-related kinase,
arginine/serine-rich [EC:2.7.11.22]
Length=730
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 31/47 (65%), Gaps = 9/47 (19%)
Query 33 REIKILQNLFGGPNIIKLLDVVKDPLS-----RTPA---LIFEYVNN 71
REIKIL+ L NI++L+D+V D +S RT A L+FEYV++
Sbjct 357 REIKILRQLHH-KNIVRLMDIVIDDISMDELKRTRANFYLVFEYVDH 402
> xla:779225 mapkapk3, MGC154360; mitogen-activated protein kinase-activated
protein kinase 3 (EC:2.7.11.1); K04444 mitogen-activated
protein kinase-activated protein kinase 3 [EC:2.7.11.1]
Length=302
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 15 GQRCVIKVLKPVKKKKIRREIKILQNLFGGPNIIKLLDVVKD 56
GQ+C +K+L K RRE++ GGP+I+ +LDV ++
Sbjct 59 GQKCALKIL--YDSPKARREVECHIRASGGPHIVHVLDVYEN 98
Lambda K H
0.323 0.141 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017039696
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40