bitscore colors: <40, 40-50 , 50-80, 80-200, >200
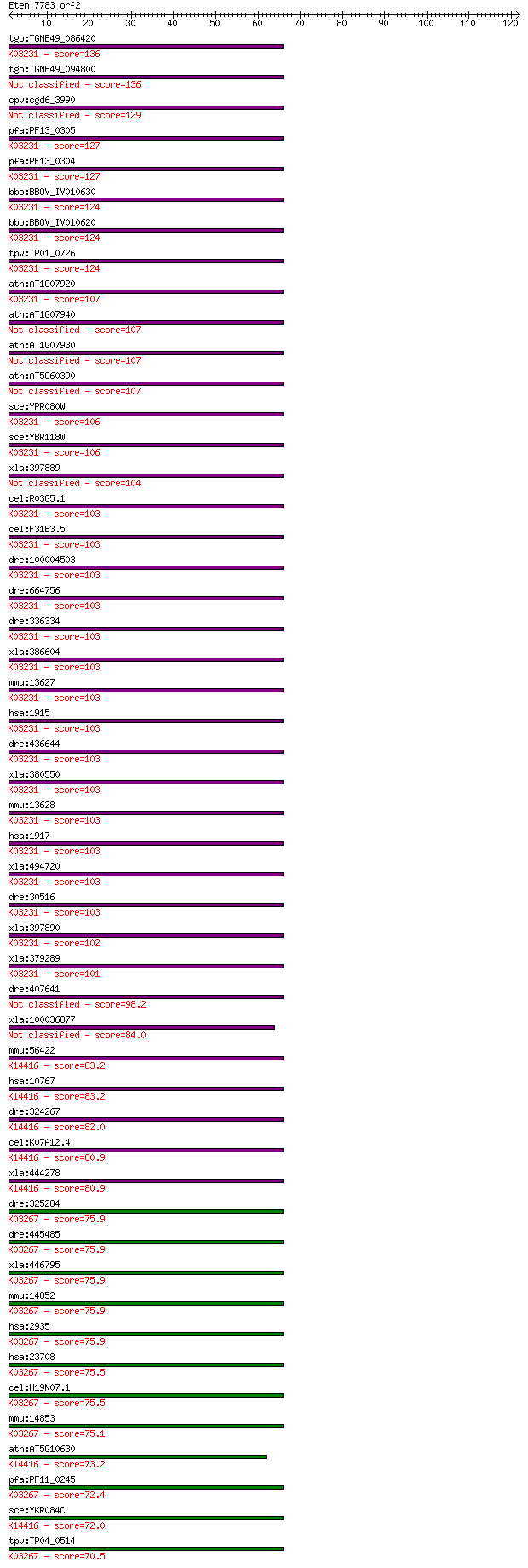
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7783_orf2
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_086420 elongation factor 1-alpha, putative (EC:2.7.... 136 1e-32
tgo:TGME49_094800 elongation factor 1-alpha, putative (EC:2.7.... 136 1e-32
cpv:cgd6_3990 elongation factor 1 alpha 129 2e-30
pfa:PF13_0305 elongation factor-1 alpha; K03231 elongation fac... 127 7e-30
pfa:PF13_0304 elongation factor-1 alpha; K03231 elongation fac... 127 7e-30
bbo:BBOV_IV010630 23.m06438; elongation factor 1-alpha (EC:3.6... 124 7e-29
bbo:BBOV_IV010620 23.m06453; elongation factor 1-alpha; K03231... 124 7e-29
tpv:TP01_0726 elongation factor 1 alpha; K03231 elongation fac... 124 8e-29
ath:AT1G07920 elongation factor 1-alpha / EF-1-alpha; K03231 e... 107 1e-23
ath:AT1G07940 elongation factor 1-alpha / EF-1-alpha 107 1e-23
ath:AT1G07930 elongation factor 1-alpha / EF-1-alpha 107 1e-23
ath:AT5G60390 elongation factor 1-alpha / EF-1-alpha 107 1e-23
sce:YPR080W TEF1; Tef1p; K03231 elongation factor 1-alpha 106 1e-23
sce:YBR118W TEF2; Tef2p; K03231 elongation factor 1-alpha 106 1e-23
xla:397889 42Sp50; elongation factor 1-alpha 104 8e-23
cel:R03G5.1 eft-4; Elongation FacTor family member (eft-4); K0... 103 1e-22
cel:F31E3.5 eft-3; Elongation FacTor family member (eft-3); K0... 103 1e-22
dre:100004503 eef1a1, MGC192680, wu:fj34g08, zgc:110335; eukar... 103 1e-22
dre:664756 MGC109885, wu:fi48f03; zgc:109885; K03231 elongatio... 103 1e-22
dre:336334 fj64c02, wu:fj64c02; zgc:73138; K03231 elongation f... 103 1e-22
xla:386604 eef1a1, EF-1-ALPHA-S, eef1a-s, eef1as; eukaryotic t... 103 1e-22
mmu:13627 Eef1a1, MGC102592, MGC103271, MGC118397, MGC18758, M... 103 1e-22
hsa:1915 EEF1A1, CCS-3, CCS3, EE1A1, EEF-1, EEF1A, EF-Tu, EF1A... 103 1e-22
dre:436644 zgc:92085; K03231 elongation factor 1-alpha 103 2e-22
xla:380550 eef1a2, MGC64523; eukaryotic translation elongation... 103 2e-22
mmu:13628 Eef1a2, Eef1a, S1, wasted, wst; eukaryotic translati... 103 2e-22
hsa:1917 EEF1A2, EEF1AL, EF-1-alpha-2, EF1A, FLJ41696, HS1, ST... 103 2e-22
xla:494720 hypothetical LOC494720; K03231 elongation factor 1-... 103 2e-22
dre:30516 ef1a, EFL1-alpha, chunp6927, eef1a, ik:tdsubc_2a3, i... 103 2e-22
xla:397890 eef1a1o, MGC86322, eef1ao, ef-1ao, xef1ao; eukaryot... 102 2e-22
xla:379289 eef1a-o1, 42Sp48, EF-1-ALPHA-O1, EF-1-alpha-O, MGC5... 101 6e-22
dre:407641 fi12b10, wu:fi12b10; si:dkey-37o8.1 98.2 5e-21
xla:100036877 hypothetical protein LOC100036877 84.0 1e-16
mmu:56422 Hbs1l, 2810035F15Rik, AI326327, eRFS; Hbs1-like (S. ... 83.2 2e-16
hsa:10767 HBS1L, DKFZp686L13262, EF-1a, ERFS, HBS1, HSPC276; H... 83.2 2e-16
dre:324267 hbs1l, wu:fc23c07, zgc:55400; HBS1-like (S. cerevis... 82.0 4e-16
cel:K07A12.4 hypothetical protein; K14416 elongation factor 1 ... 80.9 1e-15
xla:444278 hbs1l, MGC80911; HBS1-like; K14416 elongation facto... 80.9 1e-15
dre:325284 gspt1l, gspt1, hm:zehn1143, wu:fa91e05, wu:fc64f03,... 75.9 3e-14
dre:445485 gspt1, zgc:91975; G1 to S phase transition 1; K0326... 75.9 3e-14
xla:446795 gspt1, MGC80483, eRF3a, etf3a, gspt2, gst1; G1 to S... 75.9 3e-14
mmu:14852 Gspt1, AI314175, AI326449, AV307676, C79774, G1st, G... 75.9 3e-14
hsa:2935 GSPT1, 551G9.2, ETF3A, FLJ38048, FLJ39067, GST1, eRF3... 75.9 3e-14
hsa:23708 GSPT2, ERF3B, FLJ10441, GST2; G1 to S phase transiti... 75.5 4e-14
cel:H19N07.1 hypothetical protein; K03267 peptide chain releas... 75.5 4e-14
mmu:14853 Gspt2, MGC143748, MGC143749; G1 to S phase transitio... 75.1 5e-14
ath:AT5G10630 elongation factor 1-alpha, putative / EF-1-alpha... 73.2 2e-13
pfa:PF11_0245 translation elongation factor EF-1, subunit alph... 72.4 4e-13
sce:YKR084C HBS1; GTP binding protein with sequence similarity... 72.0 4e-13
tpv:TP04_0514 translation elongation factor EF-1 subunit alpha... 70.5 1e-12
> tgo:TGME49_086420 elongation factor 1-alpha, putative (EC:2.7.7.4);
K03231 elongation factor 1-alpha
Length=448
Score = 136 bits (343), Expect = 1e-32, Method: Composition-based stats.
Identities = 61/65 (93%), Positives = 63/65 (96%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALWQFETP +HYTVIDAPGHRDFIKNMITGTSQADVALLVVPA+ GGFEGA
Sbjct 67 RERGITIDIALWQFETPKYHYTVIDAPGHRDFIKNMITGTSQADVALLVVPAEAGGFEGA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> tgo:TGME49_094800 elongation factor 1-alpha, putative (EC:2.7.7.4)
Length=448
Score = 136 bits (343), Expect = 1e-32, Method: Composition-based stats.
Identities = 61/65 (93%), Positives = 63/65 (96%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALWQFETP +HYTVIDAPGHRDFIKNMITGTSQADVALLVVPA+ GGFEGA
Sbjct 67 RERGITIDIALWQFETPKYHYTVIDAPGHRDFIKNMITGTSQADVALLVVPAEAGGFEGA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> cpv:cgd6_3990 elongation factor 1 alpha
Length=435
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 60/65 (92%), Positives = 62/65 (95%), Gaps = 2/65 (3%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALWQFETP +HYTVIDAPGHRDFIKNMITGTSQADVALLVVPAD+ FEGA
Sbjct 67 RERGITIDIALWQFETPKYHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADR--FEGA 124
Query 61 FSKEG 65
FSKEG
Sbjct 125 FSKEG 129
> pfa:PF13_0305 elongation factor-1 alpha; K03231 elongation factor
1-alpha
Length=443
Score = 127 bits (320), Expect = 7e-30, Method: Composition-based stats.
Identities = 57/65 (87%), Positives = 62/65 (95%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FETP + +TVIDAPGH+DFIKNMITGTSQADVALLVVPA+ GGFEGA
Sbjct 67 RERGITIDIALWKFETPRYFFTVIDAPGHKDFIKNMITGTSQADVALLVVPAEVGGFEGA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> pfa:PF13_0304 elongation factor-1 alpha; K03231 elongation factor
1-alpha
Length=443
Score = 127 bits (320), Expect = 7e-30, Method: Composition-based stats.
Identities = 57/65 (87%), Positives = 62/65 (95%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FETP + +TVIDAPGH+DFIKNMITGTSQADVALLVVPA+ GGFEGA
Sbjct 67 RERGITIDIALWKFETPRYFFTVIDAPGHKDFIKNMITGTSQADVALLVVPAEVGGFEGA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> bbo:BBOV_IV010630 23.m06438; elongation factor 1-alpha (EC:3.6.5.3);
K03231 elongation factor 1-alpha
Length=448
Score = 124 bits (311), Expect = 7e-29, Method: Composition-based stats.
Identities = 55/65 (84%), Positives = 60/65 (92%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI LW+FET ++YTVIDAPGHRDFIKNMITGTSQADVA+LVVPA+ GGFE A
Sbjct 67 RERGITIDITLWKFETTKYYYTVIDAPGHRDFIKNMITGTSQADVAMLVVPAEAGGFEAA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> bbo:BBOV_IV010620 23.m06453; elongation factor 1-alpha; K03231
elongation factor 1-alpha
Length=448
Score = 124 bits (311), Expect = 7e-29, Method: Composition-based stats.
Identities = 55/65 (84%), Positives = 60/65 (92%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI LW+FET ++YTVIDAPGHRDFIKNMITGTSQADVA+LVVPA+ GGFE A
Sbjct 67 RERGITIDITLWKFETTKYYYTVIDAPGHRDFIKNMITGTSQADVAMLVVPAEAGGFEAA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> tpv:TP01_0726 elongation factor 1 alpha; K03231 elongation factor
1-alpha
Length=448
Score = 124 bits (311), Expect = 8e-29, Method: Composition-based stats.
Identities = 55/65 (84%), Positives = 60/65 (92%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI LW+FET ++YTVIDAPGHRDFIKNMITGTSQADVA+LVVPA+ GGFE A
Sbjct 67 RERGITIDITLWKFETGKYYYTVIDAPGHRDFIKNMITGTSQADVAMLVVPAESGGFEAA 126
Query 61 FSKEG 65
FSKEG
Sbjct 127 FSKEG 131
> ath:AT1G07920 elongation factor 1-alpha / EF-1-alpha; K03231
elongation factor 1-alpha
Length=449
Score = 107 bits (267), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 55/65 (84%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ TVIDAPGHRDFIKNMITGTSQAD A+L++ + GGFE
Sbjct 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> ath:AT1G07940 elongation factor 1-alpha / EF-1-alpha
Length=449
Score = 107 bits (267), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 55/65 (84%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ TVIDAPGHRDFIKNMITGTSQAD A+L++ + GGFE
Sbjct 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> ath:AT1G07930 elongation factor 1-alpha / EF-1-alpha
Length=372
Score = 107 bits (267), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 55/65 (84%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ TVIDAPGHRDFIKNMITGTSQAD A+L++ + GGFE
Sbjct 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> ath:AT5G60390 elongation factor 1-alpha / EF-1-alpha
Length=400
Score = 107 bits (267), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 55/65 (84%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ TVIDAPGHRDFIKNMITGTSQAD A+L++ + GGFE
Sbjct 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> sce:YPR080W TEF1; Tef1p; K03231 elongation factor 1-alpha
Length=458
Score = 106 bits (265), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FETP + TVIDAPGHRDFIKNMITGTSQAD A+L++ G FE
Sbjct 67 RERGITIDIALWKFETPKYQVTVIDAPGHRDFIKNMITGTSQADCAILIIAGGVGEFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> sce:YBR118W TEF2; Tef2p; K03231 elongation factor 1-alpha
Length=458
Score = 106 bits (265), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FETP + TVIDAPGHRDFIKNMITGTSQAD A+L++ G FE
Sbjct 67 RERGITIDIALWKFETPKYQVTVIDAPGHRDFIKNMITGTSQADCAILIIAGGVGEFEAG 126
Query 61 FSKEG 65
SK+G
Sbjct 127 ISKDG 131
> xla:397889 42Sp50; elongation factor 1-alpha
Length=463
Score = 104 bits (259), Expect = 8e-23, Method: Composition-based stats.
Identities = 48/65 (73%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+F+T F T+IDAPGHRDFIKNMITGTSQADVALLVV A G FE
Sbjct 70 RERGITIDISLWKFQTNRFTITIIDAPGHRDFIKNMITGTSQADVALLVVSAATGEFEAG 129
Query 61 FSKEG 65
S+ G
Sbjct 130 VSRNG 134
> cel:R03G5.1 eft-4; Elongation FacTor family member (eft-4);
K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (258), Expect = 1e-22, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+LVV G FE
Sbjct 67 RERGITIDIALWKFETAKYYITIIDAPGHRDFIKNMITGTSQADCAVLVVACGTGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> cel:F31E3.5 eft-3; Elongation FacTor family member (eft-3);
K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (258), Expect = 1e-22, Method: Composition-based stats.
Identities = 47/65 (72%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+LVV G FE
Sbjct 67 RERGITIDIALWKFETAKYYITIIDAPGHRDFIKNMITGTSQADCAVLVVACGTGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> dre:100004503 eef1a1, MGC192680, wu:fj34g08, zgc:110335; eukaryotic
translation elongation factor 1 alpha 1; K03231 elongation
factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> dre:664756 MGC109885, wu:fi48f03; zgc:109885; K03231 elongation
factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> dre:336334 fj64c02, wu:fj64c02; zgc:73138; K03231 elongation
factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> xla:386604 eef1a1, EF-1-ALPHA-S, eef1a-s, eef1as; eukaryotic
translation elongation factor 1 alpha 1; K03231 elongation
factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> mmu:13627 Eef1a1, MGC102592, MGC103271, MGC118397, MGC18758,
MGC27859, MGC7551, MGC8115, MGC8209; eukaryotic translation
elongation factor 1 alpha 1; K03231 elongation factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> hsa:1915 EEF1A1, CCS-3, CCS3, EE1A1, EEF-1, EEF1A, EF-Tu, EF1A,
FLJ25721, GRAF-1EF, HNGC:16303, LENG7, MGC102687, MGC131894,
MGC16224, PTI1, eEF1A-1; eukaryotic translation elongation
factor 1 alpha 1; K03231 elongation factor 1-alpha
Length=462
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> dre:436644 zgc:92085; K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> xla:380550 eef1a2, MGC64523; eukaryotic translation elongation
factor 1 alpha 2; K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> mmu:13628 Eef1a2, Eef1a, S1, wasted, wst; eukaryotic translation
elongation factor 1 alpha 2; K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> hsa:1917 EEF1A2, EEF1AL, EF-1-alpha-2, EF1A, FLJ41696, HS1,
STN, STNL; eukaryotic translation elongation factor 1 alpha
2; K03231 elongation factor 1-alpha
Length=463
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDISLWKFETTKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> xla:494720 hypothetical LOC494720; K03231 elongation factor
1-alpha
Length=461
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 49/65 (75%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET TVIDAPGHRDFIKNMITGTSQAD A+L+V A G FE
Sbjct 67 RERGITIDIALWKFETTKKVVTVIDAPGHRDFIKNMITGTSQADCAVLIVAAGTGEFEAG 126
Query 61 FSKEG 65
SKEG
Sbjct 127 ISKEG 131
> dre:30516 ef1a, EFL1-alpha, chunp6927, eef1a, ik:tdsubc_2a3,
ik:tdsubc_2b3, tdsubc_2a3, wu:fa91c07, wu:fa94b03, wu:fi13b09,
xx:tdsubc_2a3, xx:tdsubc_2b3; elongation factor 1-alpha;
K03231 elongation factor 1-alpha
Length=462
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDIALW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V G FE
Sbjct 67 RERGITIDIALWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAGGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> xla:397890 eef1a1o, MGC86322, eef1ao, ef-1ao, xef1ao; eukaryotic
translation elongation factor 1 alpha 1, oocyte form; K03231
elongation factor 1-alpha
Length=461
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/65 (70%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET F+ T+IDAPGHRDFIKNMITGTSQAD A+L+V G FE
Sbjct 67 RERGITIDISLWKFETGKFYITIIDAPGHRDFIKNMITGTSQADCAVLIVAGGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> xla:379289 eef1a-o1, 42Sp48, EF-1-ALPHA-O1, EF-1-alpha-O, MGC53846;
elongation factor 1 alpha, oocyte form; K03231 elongation
factor 1-alpha
Length=461
Score = 101 bits (251), Expect = 6e-22, Method: Composition-based stats.
Identities = 45/65 (69%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+LW+FET ++ T+IDAPGHRDFIKNMITGTSQAD A+L+V G FE
Sbjct 67 RERGITIDISLWKFETGKYYITIIDAPGHRDFIKNMITGTSQADCAVLIVAGGVGEFEAG 126
Query 61 FSKEG 65
SK G
Sbjct 127 ISKNG 131
> dre:407641 fi12b10, wu:fi12b10; si:dkey-37o8.1
Length=458
Score = 98.2 bits (243), Expect = 5e-21, Method: Composition-based stats.
Identities = 44/65 (67%), Positives = 52/65 (80%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGITIDI+L +F T + +T+IDAPGHRDFIKNMITGTSQAD ALL+V A +G FE
Sbjct 67 RERGITIDISLLKFNTQKYTFTIIDAPGHRDFIKNMITGTSQADAALLIVSAAKGEFEAG 126
Query 61 FSKEG 65
S+ G
Sbjct 127 ISRNG 131
> xla:100036877 hypothetical protein LOC100036877
Length=462
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 43/63 (68%), Positives = 48/63 (76%), Gaps = 1/63 (1%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG+TI +F T HYTVIDAPGHRDFIKNMITG SQADVALL++PAD G F A
Sbjct 67 RERGVTIACTTKEFFTTTKHYTVIDAPGHRDFIKNMITGASQADVALLMIPAD-GNFTAA 125
Query 61 FSK 63
+K
Sbjct 126 IAK 128
> mmu:56422 Hbs1l, 2810035F15Rik, AI326327, eRFS; Hbs1-like (S.
cerevisiae); K14416 elongation factor 1 alpha-like protein
Length=679
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 48/65 (73%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG+T+D+ + +FET T++DAPGH+DFI NMITG +QADVA+LVV A +G FE
Sbjct 315 RERGVTMDVGMTKFETTTKVITLMDAPGHKDFIPNMITGAAQADVAVLVVDASRGEFEAG 374
Query 61 FSKEG 65
F G
Sbjct 375 FETGG 379
> hsa:10767 HBS1L, DKFZp686L13262, EF-1a, ERFS, HBS1, HSPC276;
HBS1-like (S. cerevisiae); K14416 elongation factor 1 alpha-like
protein
Length=642
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 48/65 (73%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG+T+D+ + +FET T++DAPGH+DFI NMITG +QADVA+LVV A +G FE
Sbjct 278 RERGVTMDVGMTKFETTTKVITLMDAPGHKDFIPNMITGAAQADVAVLVVDASRGEFEAG 337
Query 61 FSKEG 65
F G
Sbjct 338 FETGG 342
> dre:324267 hbs1l, wu:fc23c07, zgc:55400; HBS1-like (S. cerevisiae);
K14416 elongation factor 1 alpha-like protein
Length=653
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 36/65 (55%), Positives = 49/65 (75%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R+RG+T+D+ + +FET + T++DAPGH+DFI NMITG +QADVA+LVV A +G FE
Sbjct 318 RDRGVTMDVGMTKFETDSKVVTLMDAPGHKDFIPNMITGAAQADVAVLVVDASRGEFEAG 377
Query 61 FSKEG 65
F G
Sbjct 378 FEAGG 382
> cel:K07A12.4 hypothetical protein; K14416 elongation factor
1 alpha-like protein
Length=592
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/65 (58%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG+T+DI FET ++DAPGH+DFI NMITGTSQAD A+LVV A G FE
Sbjct 230 RERGVTMDIGRTSFETSHRRIVLLDAPGHKDFISNMITGTSQADAAILVVNATTGEFETG 289
Query 61 FSKEG 65
F G
Sbjct 290 FENGG 294
> xla:444278 hbs1l, MGC80911; HBS1-like; K14416 elongation factor
1 alpha-like protein
Length=678
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 35/65 (53%), Positives = 48/65 (73%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R+RG+T+D+ + +FET + T++DAPGH+DFI NMITG +QADVA+L V A +G FE
Sbjct 314 RQRGVTMDVGMTKFETKSKVITLMDAPGHKDFIPNMITGAAQADVAVLAVDASRGEFEAG 373
Query 61 FSKEG 65
F G
Sbjct 374 FEAGG 378
> dre:325284 gspt1l, gspt1, hm:zehn1143, wu:fa91e05, wu:fc64f03,
zehn1143; G1 to S phase transition 1, like; K03267 peptide
chain release factor subunit 3
Length=577
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 212 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 271
Query 61 FSKEG 65
F K G
Sbjct 272 FEKGG 276
> dre:445485 gspt1, zgc:91975; G1 to S phase transition 1; K03267
peptide chain release factor subunit 3
Length=564
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 199 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 258
Query 61 FSKEG 65
F K G
Sbjct 259 FEKGG 263
> xla:446795 gspt1, MGC80483, eRF3a, etf3a, gspt2, gst1; G1 to
S phase transition 1; K03267 peptide chain release factor subunit
3
Length=553
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 188 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 247
Query 61 FSKEG 65
F K G
Sbjct 248 FEKGG 252
> mmu:14852 Gspt1, AI314175, AI326449, AV307676, C79774, G1st,
Gst-1, MGC36230, MGC36735; G1 to S phase transition 1; K03267
peptide chain release factor subunit 3
Length=635
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 270 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 329
Query 61 FSKEG 65
F K G
Sbjct 330 FEKGG 334
> hsa:2935 GSPT1, 551G9.2, ETF3A, FLJ38048, FLJ39067, GST1, eRF3a;
G1 to S phase transition 1; K03267 peptide chain release
factor subunit 3
Length=636
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 271 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 330
Query 61 FSKEG 65
F K G
Sbjct 331 FEKGG 335
> hsa:23708 GSPT2, ERF3B, FLJ10441, GST2; G1 to S phase transition
2; K03267 peptide chain release factor subunit 3
Length=628
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 263 RDKGKTVEVGRAYFETERKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 322
Query 61 FSKEG 65
F K G
Sbjct 323 FEKGG 327
> cel:H19N07.1 hypothetical protein; K03267 peptide chain release
factor subunit 3
Length=532
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RE+G T+++ FET H+T++DAPGH+ F+ NMI G +QAD+A+LV+ A +G FE
Sbjct 169 REKGKTVEVGRAYFETEKRHFTILDAPGHKSFVPNMIVGANQADLAVLVISARRGEFETG 228
Query 61 FSKEG 65
F + G
Sbjct 229 FDRGG 233
> mmu:14853 Gspt2, MGC143748, MGC143749; G1 to S phase transition
2; K03267 peptide chain release factor subunit 3
Length=632
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET H+T++DAPGH+ F+ NMI G SQAD+A+LV+ A +G FE
Sbjct 267 RDKGKTVEVGRAYFETEKKHFTILDAPGHKSFVPNMIGGASQADLAVLVISARKGEFETG 326
Query 61 FSKEG 65
F K G
Sbjct 327 FEKGG 331
> ath:AT5G10630 elongation factor 1-alpha, putative / EF-1-alpha,
putative; K14416 elongation factor 1 alpha-like protein
Length=667
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 30/61 (49%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERGIT+ +A+ F + H ++D+PGH+DF+ NMI G +QAD A+LV+ A G FE
Sbjct 299 RERGITMTVAVAYFNSKRHHVVLLDSPGHKDFVPNMIAGATQADAAILVIDASVGAFEAG 358
Query 61 F 61
F
Sbjct 359 F 359
> pfa:PF11_0245 translation elongation factor EF-1, subunit alpha,
putative; K03267 peptide chain release factor subunit 3
Length=555
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
R++G T+++ FET +T++DAPGH++FI NMI+G +QAD+ +L++ A +G FE
Sbjct 179 RQKGKTVEVGRAHFETKDRRFTILDAPGHKNFIPNMISGAAQADIGVLIISARKGEFETG 238
Query 61 FSKEG 65
F + G
Sbjct 239 FERGG 243
> sce:YKR084C HBS1; GTP binding protein with sequence similarity
to the elongation factor class of G proteins, EF-1alpha and
Sup35p; associates with Dom34p, and shares a similar genetic
relationship with genes that encode ribosomal protein components;
K14416 elongation factor 1 alpha-like protein
Length=611
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 41/65 (63%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG+T+ I F T ++T++DAPGHRDF+ N I G SQAD+A+L V FE
Sbjct 227 RERGVTVSICTSHFSTHRANFTIVDAPGHRDFVPNAIMGISQADMAILCVDCSTNAFESG 286
Query 61 FSKEG 65
F +G
Sbjct 287 FDLDG 291
> tpv:TP04_0514 translation elongation factor EF-1 subunit alpha;
K03267 peptide chain release factor subunit 3
Length=539
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 1 RERGITIDIALWQFETPAFHYTVIDAPGHRDFIKNMITGTSQADVALLVVPADQGGFEGA 60
RERG TI++ + ET +T++DAPGHR+++ NMI G +QAD +L++ A +G FE
Sbjct 162 RERGKTIEVGRAKIETKNRRFTILDAPGHRNYVPNMIDGATQADCGVLIISARKGEFETG 221
Query 61 FSKEG 65
F + G
Sbjct 222 FERGG 226
Lambda K H
0.326 0.141 0.511
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40