bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7776_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
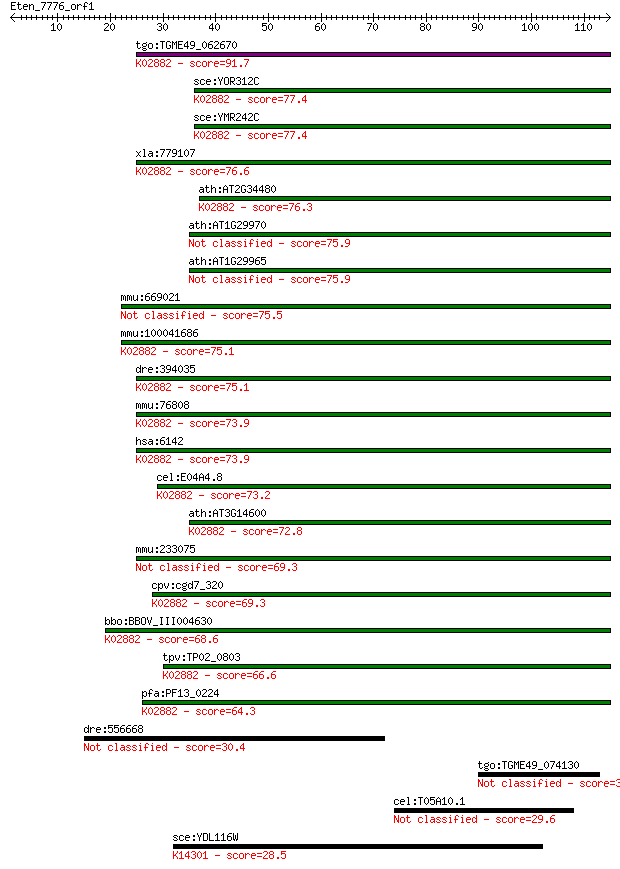
tgo:TGME49_062670 60S ribosomal protein L18a, putative ; K0288... 91.7 5e-19
sce:YOR312C RPL20B, RPL18A1; L20B; L18B; K02882 large subunit ... 77.4 9e-15
sce:YMR242C RPL20A, RPL18A2; L20A; L18A; K02882 large subunit ... 77.4 9e-15
xla:779107 rpl18a, MGC64263; ribosomal protein L18a; K02882 la... 76.6 2e-14
ath:AT2G34480 60S ribosomal protein L18A (RPL18aB); K02882 lar... 76.3 2e-14
ath:AT1G29970 RPL18AA (60S RIBOSOMAL PROTEIN L18A-1); structur... 75.9 3e-14
ath:AT1G29965 60S ribosomal protein L18A (RPL18aA) 75.9 3e-14
mmu:669021 60S ribosomal protein L18a-like 75.5 4e-14
mmu:100041686 Gm15427, OTTMUSG00000021609; predicted pseudogen... 75.1 6e-14
dre:394035 rpl18a, MGC56546, rpl-20Co, zgc:56546; ribosomal pr... 75.1 6e-14
mmu:76808 Rpl18a, 2510019J09Rik; ribosomal protein L18A; K0288... 73.9 1e-13
hsa:6142 RPL18A; ribosomal protein L18a; K02882 large subunit ... 73.9 1e-13
cel:E04A4.8 rpl-20; Ribosomal Protein, Large subunit family me... 73.2 2e-13
ath:AT3G14600 60S ribosomal protein L18A (RPL18aC); K02882 lar... 72.8 3e-13
mmu:233075 Gm4883, EG233075; predicted gene 4883 69.3
cpv:cgd7_320 60S ribosomal protein L18A ; K02882 large subunit... 69.3 3e-12
bbo:BBOV_III004630 17.m07414; 60S ribosomal protein L18ae; K02... 68.6 5e-12
tpv:TP02_0803 60S ribosomal protein L18; K02882 large subunit ... 66.6 2e-11
pfa:PF13_0224 60S ribosomal protein L18, putative; K02882 larg... 64.3 9e-11
dre:556668 MGC84409 protein-like 30.4 1.4
tgo:TGME49_074130 hypothetical protein 30.0 2.1
cel:T05A10.1 sma-9; SMAll family member (sma-9) 29.6 2.2
sce:YDL116W NUP84; Nup84p; K14301 nuclear pore complex protein... 28.5 5.7
> tgo:TGME49_062670 60S ribosomal protein L18a, putative ; K02882
large subunit ribosomal protein L18Ae
Length=183
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 51/90 (56%), Positives = 67/90 (74%), Gaps = 4/90 (4%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKAD TLQQ I Q+QVVGR++P A + +R+RLFA N+VLAVS+FWYLL++++
Sbjct 1 MKADPTLQQKISQYQVVGRKQPTEAEPNPSL----FRMRLFARNKVLAVSKFWYLLKKMK 56
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K++ GE+L V EI KR T KNFGVWL
Sbjct 57 KVKKSTGEILAVNEIREKRPTFVKNFGVWL 86
> sce:YOR312C RPL20B, RPL18A1; L20B; L18B; K02882 large subunit
ribosomal protein L18Ae
Length=172
Score = 77.4 bits (189), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 54/79 (68%), Gaps = 4/79 (5%)
Query 36 RQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVLE 95
+++QV+GR+ P + +R+R+FA+NEV+A SR+WY L++L K+K+A GE++
Sbjct 5 KEYQVIGRRLPTESVPEPKL----FRMRIFASNEVIAKSRYWYFLQKLHKVKKASGEIVS 60
Query 96 VVEINPKRATNPKNFGVWL 114
+ +IN T KNFGVW+
Sbjct 61 INQINEAHPTKVKNFGVWV 79
> sce:YMR242C RPL20A, RPL18A2; L20A; L18A; K02882 large subunit
ribosomal protein L18Ae
Length=172
Score = 77.4 bits (189), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 54/79 (68%), Gaps = 4/79 (5%)
Query 36 RQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVLE 95
+++QV+GR+ P + +R+R+FA+NEV+A SR+WY L++L K+K+A GE++
Sbjct 5 KEYQVIGRRLPTESVPEPKL----FRMRIFASNEVIAKSRYWYFLQKLHKVKKASGEIVS 60
Query 96 VVEINPKRATNPKNFGVWL 114
+ +IN T KNFGVW+
Sbjct 61 INQINEAHPTKVKNFGVWV 79
> xla:779107 rpl18a, MGC64263; ribosomal protein L18a; K02882
large subunit ribosomal protein L18Ae
Length=176
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 56/90 (62%), Gaps = 8/90 (8%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL R+++V+GR P A + P YR+R+FA N V+A SRFWY + QL+
Sbjct 1 MKASGTL----REYKVIGRSLPTPKAPSP----PLYRMRIFAPNHVVAKSRFWYFVSQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K+A GE++ ++ K KNFG+WL
Sbjct 53 KMKKASGEIVYCGQVFEKTPLKVKNFGIWL 82
> ath:AT2G34480 60S ribosomal protein L18A (RPL18aB); K02882 large
subunit ribosomal protein L18Ae
Length=178
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/78 (46%), Positives = 51/78 (65%), Gaps = 4/78 (5%)
Query 37 QFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVLEV 96
Q+QVVGR P YR++L+A NEV A S+FWY LR+L+K+K+++G++L +
Sbjct 8 QYQVVGRALPTEKDVQPKI----YRMKLWATNEVRAKSKFWYFLRKLKKVKKSNGQMLAI 63
Query 97 VEINPKRATNPKNFGVWL 114
EI K T KNFG+WL
Sbjct 64 NEIYEKNPTTIKNFGIWL 81
> ath:AT1G29970 RPL18AA (60S RIBOSOMAL PROTEIN L18A-1); structural
constituent of ribosome
Length=312
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 52/80 (65%), Gaps = 4/80 (5%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
+ Q+QVVGR P YR++L+A NEVLA S+FWY LR+ +K+K+++G++L
Sbjct 140 LHQYQVVGRALPTEKDEQPKI----YRMKLWATNEVLAKSKFWYYLRRQKKVKKSNGQML 195
Query 95 EVVEINPKRATNPKNFGVWL 114
+ EI K T KNFG+WL
Sbjct 196 AINEIFEKNPTTIKNFGIWL 215
> ath:AT1G29965 60S ribosomal protein L18A (RPL18aA)
Length=178
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 52/80 (65%), Gaps = 4/80 (5%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
+ Q+QVVGR P YR++L+A NEVLA S+FWY LR+ +K+K+++G++L
Sbjct 6 LHQYQVVGRALPTEKDEQPKI----YRMKLWATNEVLAKSKFWYYLRRQKKVKKSNGQML 61
Query 95 EVVEINPKRATNPKNFGVWL 114
+ EI K T KNFG+WL
Sbjct 62 AINEIFEKNPTTIKNFGIWL 81
> mmu:669021 60S ribosomal protein L18a-like
Length=217
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 41/98 (41%), Positives = 57/98 (58%), Gaps = 13/98 (13%)
Query 22 GGE-----MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRF 76
GGE MKA TL R+++VVGR P P YR+R+FA N V+A SRF
Sbjct 34 GGERACSAMKASGTL----REYKVVGRCLPTPKCHTP----PLYRMRIFAPNHVVAKSRF 85
Query 77 WYLLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
WY + QL+K+K++ GE++ ++ K KNFG+WL
Sbjct 86 WYFVSQLKKMKKSSGEIVYCGQVFEKSPLRVKNFGIWL 123
> mmu:100041686 Gm15427, OTTMUSG00000021609; predicted pseudogene
15427; K02882 large subunit ribosomal protein L18Ae
Length=191
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/98 (41%), Positives = 57/98 (58%), Gaps = 13/98 (13%)
Query 22 GGE-----MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRF 76
GGE MKA TL R+++VVGR P P YR+R+FA N V+A SRF
Sbjct 8 GGERVRSAMKASGTL----REYKVVGRCLPTPKCHTP----PLYRMRIFAPNHVVAKSRF 59
Query 77 WYLLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
WY + QL+K+K++ GE++ ++ K KNFG+WL
Sbjct 60 WYFVSQLKKMKKSSGEIVYCRQVFEKSPLRVKNFGIWL 97
> dre:394035 rpl18a, MGC56546, rpl-20Co, zgc:56546; ribosomal
protein L18a; K02882 large subunit ribosomal protein L18Ae
Length=176
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/90 (45%), Positives = 55/90 (61%), Gaps = 8/90 (8%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL R+++VVGR P+ A P YR+R+FA N V+A SRFWY + QLR
Sbjct 1 MKASGTL----REYKVVGRLLPSVKNPAP----PLYRMRIFAPNHVVAKSRFWYFVSQLR 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K+A GE++ + K KNFG+WL
Sbjct 53 KMKKASGEIVYCGLVFEKSPLKVKNFGIWL 82
> mmu:76808 Rpl18a, 2510019J09Rik; ribosomal protein L18A; K02882
large subunit ribosomal protein L18Ae
Length=176
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 54/90 (60%), Gaps = 8/90 (8%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL R+++VVGR P P YR+R+FA N V+A SRFWY + QL+
Sbjct 1 MKASGTL----REYKVVGRCLPTPKCHTP----PLYRMRIFAPNHVVAKSRFWYFVSQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K++ GE++ ++ K KNFG+WL
Sbjct 53 KMKKSSGEIVYCGQVFEKSPLRVKNFGIWL 82
> hsa:6142 RPL18A; ribosomal protein L18a; K02882 large subunit
ribosomal protein L18Ae
Length=176
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 54/90 (60%), Gaps = 8/90 (8%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL R+++VVGR P P YR+R+FA N V+A SRFWY + QL+
Sbjct 1 MKASGTL----REYKVVGRCLPTPKCHTP----PLYRMRIFAPNHVVAKSRFWYFVSQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K++ GE++ ++ K KNFG+WL
Sbjct 53 KMKKSSGEIVYCGQVFEKSPLRVKNFGIWL 82
> cel:E04A4.8 rpl-20; Ribosomal Protein, Large subunit family
member (rpl-20); K02882 large subunit ribosomal protein L18Ae
Length=180
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 54/86 (62%), Gaps = 4/86 (4%)
Query 29 KTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKR 88
K L + + ++ VVGR+ P TP +++++FA N V+A SRFWY + LR++K+
Sbjct 4 KALGETLNEYVVVGRKIPTEKEPV----TPIWKMQIFATNHVIAKSRFWYFVSMLRRVKK 59
Query 89 AHGEVLEVVEINPKRATNPKNFGVWL 114
A+GE+L + ++ K KN+GVWL
Sbjct 60 ANGEILSIKQVFEKNPGTVKNYGVWL 85
> ath:AT3G14600 60S ribosomal protein L18A (RPL18aC); K02882 large
subunit ribosomal protein L18Ae
Length=178
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 50/80 (62%), Gaps = 4/80 (5%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
Q+QVVGR P YR++L+ NEV A S+FWY +R+L+K+K+++G++L
Sbjct 6 FHQYQVVGRALPTENDEHPKI----YRMKLWGRNEVCAKSKFWYFMRKLKKVKKSNGQML 61
Query 95 EVVEINPKRATNPKNFGVWL 114
+ EI K T KN+G+WL
Sbjct 62 AINEIFEKNPTTIKNYGIWL 81
> mmu:233075 Gm4883, EG233075; predicted gene 4883
Length=175
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 52/90 (57%), Gaps = 8/90 (8%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL + +VVGR PA P YR+R+FA N V+A S FWY + QL+
Sbjct 1 MKASGTLWE----CKVVGRCLPAPTCHTP----PLYRMRIFAPNHVVAKSHFWYFVLQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
K+K++ GE++ ++ K KNFG+WL
Sbjct 53 KMKKSSGEIVYCGKVFEKSPLRVKNFGIWL 82
> cpv:cgd7_320 60S ribosomal protein L18A ; K02882 large subunit
ribosomal protein L18Ae
Length=185
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 4/87 (4%)
Query 28 DKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIK 87
D +LQ + Q+ V+GR +P +R+ +FA NEV+A SRF+Y + +L K+K
Sbjct 6 DPSLQMKVNQYMVIGRGKPTELNPHPKV----FRVCIFAPNEVVAKSRFFYFMSRLNKVK 61
Query 88 RAHGEVLEVVEINPKRATNPKNFGVWL 114
+A+GE+L V I KR T KNF + L
Sbjct 62 KANGEILSVTRIFEKRPTYVKNFAILL 88
> bbo:BBOV_III004630 17.m07414; 60S ribosomal protein L18ae; K02882
large subunit ribosomal protein L18Ae
Length=188
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 57/97 (58%), Gaps = 8/97 (8%)
Query 19 QPLGGEMKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATP-AYRLRLFAANEVLAVSRFW 77
+P G +++ +L+Q + + VVGR AA + P YR+ +FA N VLA RFW
Sbjct 3 KPKGSQIRP--SLKQNLTHYHVVGR-----AAPSKKNPNPLIYRMNIFAKNSVLAKGRFW 55
Query 78 YLLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWL 114
Y +R+L+K KR+ GE+L +N + T KN+GV L
Sbjct 56 YFMRKLQKAKRSGGEILACNVLNEVKPTQAKNYGVLL 92
> tpv:TP02_0803 60S ribosomal protein L18; K02882 large subunit
ribosomal protein L18Ae
Length=191
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/85 (41%), Positives = 48/85 (56%), Gaps = 4/85 (4%)
Query 30 TLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRA 89
+L Q + Q+ +VGR AA YR+ +FA N VLA SRFWY +++L K KR+
Sbjct 15 SLGQNLVQYHIVGR----AAPTKKNPNPNTYRMSVFAKNSVLAKSRFWYFMKRLNKAKRS 70
Query 90 HGEVLEVVEINPKRATNPKNFGVWL 114
GE+L +N + KNFGV L
Sbjct 71 GGEILACNRLNEPKKGQVKNFGVLL 95
> pfa:PF13_0224 60S ribosomal protein L18, putative; K02882 large
subunit ribosomal protein L18Ae
Length=184
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 50/89 (56%), Gaps = 4/89 (4%)
Query 26 KADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRK 85
K D +L I Q+ +VGR P A YR+ +FA N+ A SRFWY ++++ K
Sbjct 4 KIDNSLNTNIHQYHIVGRAIPTAKDKNPNV----YRMCIFAKNDTNAKSRFWYFMKKINK 59
Query 86 IKRAHGEVLEVVEINPKRATNPKNFGVWL 114
+K+++GE+L +I + KN+GV L
Sbjct 60 LKKSNGELLACEQIKERFPLRVKNYGVLL 88
> dre:556668 MGC84409 protein-like
Length=386
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 15 VCTLQPLGGEMKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVL 71
V +P G +KAD+ P+ + + +P A +A P Y + AA++++
Sbjct 265 VAAAEPAGNLLKADQNPTSPVGKRPTSSKFQPVAQETCSACLKPVYPMERMAADKLI 321
> tgo:TGME49_074130 hypothetical protein
Length=1904
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 90 HGEVLEVVEINPKRATNPKNFGV 112
HGEV E+ + NP+ NP NF V
Sbjct 1018 HGEVREIYQGNPQLDENPVNFSV 1040
> cel:T05A10.1 sma-9; SMAll family member (sma-9)
Length=2173
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 74 SRFWYLLRQLRKIKRAHGEVLEVVEINPKRATNP 107
+RF Y +R+ +KI + ++EV++ +P A P
Sbjct 864 ARFVYQMREFQKINNMNDRLIEVLKTDPAAAATP 897
> sce:YDL116W NUP84; Nup84p; K14301 nuclear pore complex protein
Nup107
Length=726
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query 32 QQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHG 91
Q PI F ++ R AA A A + ++ + +RFW+L+ L + A
Sbjct 30 QNPIDPFNIIREFRSAAGQLALDLANSGDESNVISSKDWELEARFWHLVELLLVFRNADL 89
Query 92 EVLEVVEINP 101
+ L+ +E++P
Sbjct 90 D-LDEMELHP 98
Lambda K H
0.326 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40