bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7692_orf1
Length=101
Score E
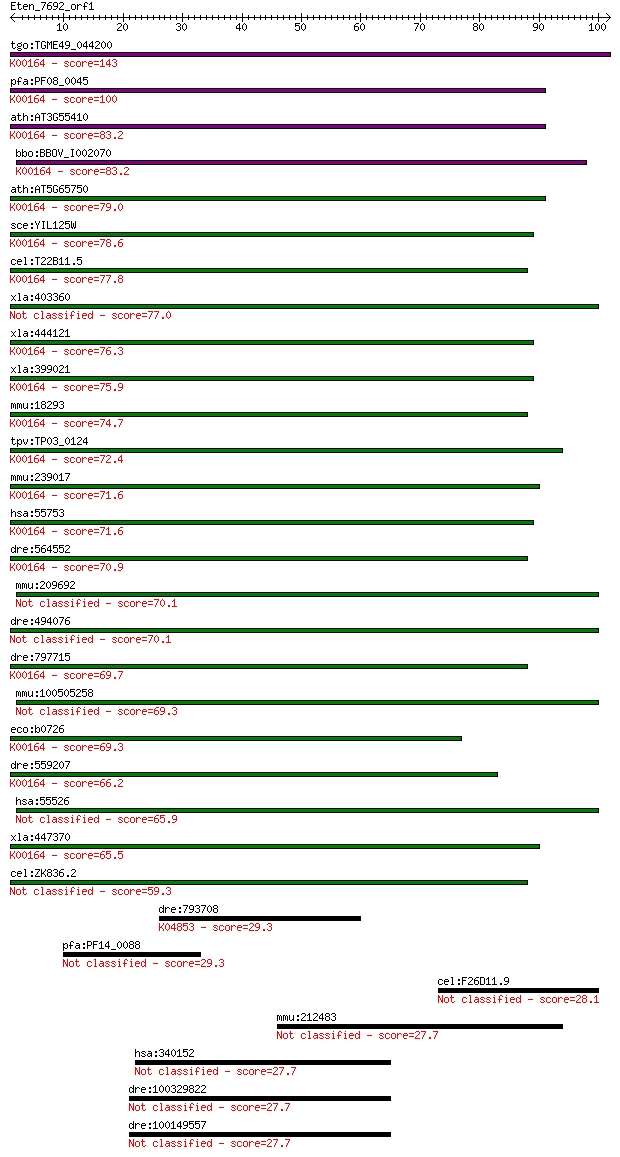
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 143 1e-34
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 100 8e-22
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 83.2 2e-16
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 83.2 2e-16
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 79.0 3e-15
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 78.6 4e-15
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 77.8 9e-15
xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolas... 77.0 1e-14
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 76.3 2e-14
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 75.9 3e-14
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 74.7 7e-14
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 72.4 4e-13
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 71.6 5e-13
hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);... 71.6 6e-13
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 70.9 9e-13
mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transke... 70.1 1e-12
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 70.1 2e-12
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 69.7 2e-12
mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 compone... 69.3 3e-12
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 69.3 3e-12
dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehyd... 66.2 2e-11
hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogena... 65.9 3e-11
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 65.5 4e-11
cel:ZK836.2 hypothetical protein 59.3 3e-09
dre:793708 calcium channel, voltage-dependent, L type, alpha 1... 29.3 2.9
pfa:PF14_0088 aldo/keto reductase, putative 29.3 3.6
cel:F26D11.9 clec-217; C-type LECtin family member (clec-217) 28.1 8.0
mmu:212483 Fam193b, IRIZIO, Kiaa1931, MGC29235, MGC38018, MGC3... 27.7 8.6
hsa:340152 ZC3H12D, C6orf95, FLJ00361, FLJ46041, MCPIP4, TFL, ... 27.7 8.8
dre:100329822 NOL1/NOP2/Sun domain family, member 6-like 27.7 9.6
dre:100149557 NOL1/NOP2/Sun domain family, member 6-like 27.7 9.6
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 66/101 (65%), Positives = 81/101 (80%), Gaps = 2/101 (1%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RLI C GQ+YYDL A +D++++ G + G+K+A+AR+EQLSPFPFDL I DL+ +PNL
Sbjct 969 RLIACSGQVYYDLIAGKDKMKN--GDENGDGDKIAIARMEQLSPFPFDLFIEDLKRFPNL 1026
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNRIERPIY 101
++VVWAQEEPMNQGAW YTSKRIES LRHL PN I PIY
Sbjct 1027 KSVVWAQEEPMNQGAWFYTSKRIESSLRHLNFPNGIRSPIY 1067
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 100 bits (250), Expect = 8e-22, Method: Composition-based stats.
Identities = 47/90 (52%), Positives = 59/90 (65%), Gaps = 9/90 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R+ILC GQ+YYDL R N + VA+AR+EQLSPFPF + DLQ YPNL
Sbjct 896 RIILCSGQVYYDLLNYR---------YTNKIDDVAIARIEQLSPFPFKQIMNDLQTYPNL 946
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHL 90
R ++WAQEE MN G W Y S+RIE+ ++ L
Sbjct 947 RDIIWAQEEHMNMGPWFYVSRRIEASIKQL 976
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/90 (47%), Positives = 60/90 (66%), Gaps = 10/90 (11%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RL+LC G++YY+L ER ++ G +D VA+ RVEQL PFP+DL R+L+ YPN
Sbjct 893 RLVLCSGKVYYELDDERKKV----GATD-----VAICRVEQLCPFPYDLIQRELKRYPNA 943
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHL 90
+VW QEE MN GA+SY S R+ + +R +
Sbjct 944 E-IVWCQEEAMNMGAFSYISPRLWTAMRSV 972
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/96 (47%), Positives = 57/96 (59%), Gaps = 9/96 (9%)
Query 2 LILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNLR 61
LILC GQ+Y+D+ DR+ SE + VA+ VEQL PFP +L+ YPNL+
Sbjct 772 LILCSGQVYFDI---SDRV-SELSVGN-----VAVTTVEQLCPFPAGALKSELERYPNLK 822
Query 62 TVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNRIE 97
+VW QEE N G WSY S RI S L HLG R+E
Sbjct 823 RLVWCQEEHANAGGWSYVSPRICSLLAHLGSGLRLE 858
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 57/90 (63%), Gaps = 10/90 (11%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RL+LC G++YY+L ER + ++ VA+ RVEQL PFP+DL R+L+ YPN
Sbjct 897 RLVLCSGKVYYELDEERKKSETK---------DVAICRVEQLCPFPYDLIQRELKRYPNA 947
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHL 90
+VW QEEPMN G + Y + R+ + ++ L
Sbjct 948 E-IVWCQEEPMNMGGYQYIALRLCTAMKAL 976
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 78.6 bits (192), Expect = 4e-15, Method: Composition-based stats.
Identities = 37/89 (41%), Positives = 51/89 (57%), Gaps = 11/89 (12%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKV-ALARVEQLSPFPFDLAIRDLQIYPN 59
RL+L GQ+Y LH R+ + G+K A ++EQL PFPF L YPN
Sbjct 892 RLVLLSGQVYTALHKRRESL----------GDKTTAFLKIEQLHPFPFAQLRDSLNSYPN 941
Query 60 LRTVVWAQEEPMNQGAWSYTSKRIESCLR 88
L +VW QEEP+N G+W+YT R+ + L+
Sbjct 942 LEEIVWCQEEPLNMGSWAYTEPRLHTTLK 970
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 77.8 bits (190), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 52/87 (59%), Gaps = 9/87 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R++ C G++YYD+ A R + E N VAL RVEQLSPFP+DL ++ + Y
Sbjct 902 RVVFCTGKVYYDMVAARKHVGKE--------NDVALVRVEQLSPFPYDLVQQECRKYQGA 953
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCL 87
++WAQEE N GAWS+ RI S L
Sbjct 954 E-ILWAQEEHKNMGAWSFVQPRINSLL 979
> xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=927
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 57/100 (57%), Gaps = 11/100 (11%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
++ILC G+ YY LH +R+ + + G A+ RVE+L PFP + +++ YP
Sbjct 812 KVILCSGKHYYALHKQREALGEQ-------GRSSAIIRVEELCPFPLEALQQEIHRYPKA 864
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNR-IERP 99
+ +W+QEEP N GAW++ + R E + L C R + RP
Sbjct 865 KDFIWSQEEPQNMGAWTFVAPRFE---KQLACKLRLVSRP 901
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/88 (45%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RLI C G++YYDL ER E VA+ RVEQLSPFPFDL +++Q YPN
Sbjct 895 RLIFCAGKVYYDLTKERSGRGME--------GDVAITRVEQLSPFPFDLVEKEVQKYPNA 946
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLR 88
+VW QEE NQG + Y R+ + +
Sbjct 947 -DLVWCQEEHKNQGYYDYVKPRLRTTIH 973
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 54/88 (61%), Gaps = 9/88 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RLI C G++YY+L ER D G+ VA+ARVEQLSPFPFDL +++Q YPN
Sbjct 898 RLIFCTGKVYYELTKER-------SGRDMEGD-VAIARVEQLSPFPFDLVEKEVQKYPNA 949
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLR 88
+VW QEE NQG + Y R+ + +
Sbjct 950 -DLVWCQEEHKNQGYYDYVKPRLRTTIH 976
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 52/87 (59%), Gaps = 9/87 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RL+ C G++YYDL ER + N +VA+ R+EQLSPFPFDL +++ Q YPN
Sbjct 900 RLLFCTGKVYYDLTRER--------KARNMEEEVAITRIEQLSPFPFDLLLKEAQKYPNA 951
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCL 87
+ W QEE NQG + Y R+ + +
Sbjct 952 E-LAWCQEEHKNQGYYDYVKPRLRTTI 977
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 12/97 (12%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R+I+C GQIYYDL EY S+ + +AR+E+++PFP + DL+ + NL
Sbjct 904 RMIMCSGQIYYDL--------LEYRDSNEEWKNIPVARIEEITPFPAQNILDDLKKFKNL 955
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRHLG----CP 93
T+VW QEE N G + + +R+ + L+ L CP
Sbjct 956 ETLVWCQEEHENSGCFYFLRERLNNVLKILKNEGHCP 992
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 38/89 (42%), Positives = 51/89 (57%), Gaps = 9/89 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RLI C G++YYDL ER +S +VA+ R+EQ+SPFPFDL +R+ + Y
Sbjct 905 RLIFCTGKVYYDLVKER--------SSQGLEQQVAITRLEQISPFPFDLIMREAEKYSGA 956
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRH 89
+VW QEE N G + Y S R + L H
Sbjct 957 E-LVWCQEEHKNMGYYDYISPRFMTLLGH 984
> hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=953
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
RLI C G++YYDL ER +S + KVA+ R+EQ+SPFPFDL ++ + YP
Sbjct 829 RLIFCTGKVYYDLVKER--------SSQDLEEKVAITRLEQISPFPFDLIKQEAEKYPGA 880
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLR 88
+ W QEE N G + Y S R + LR
Sbjct 881 E-LAWCQEEHKNMGYYDYISPRFMTILR 907
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 51/87 (58%), Gaps = 9/87 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R++ C G+IYY+L ER + N N VA+ R+EQLSPFPFDL + + +PN
Sbjct 899 RIVFCTGKIYYELTRER--------KARNMENSVAITRIEQLSPFPFDLVRAETEKFPNA 950
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCL 87
+VW QEE NQG + Y R+ + +
Sbjct 951 -DLVWCQEEHKNQGYYDYVKPRMRTTI 976
> mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=921
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 37/99 (37%), Positives = 56/99 (56%), Gaps = 11/99 (11%)
Query 2 LILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNLR 61
LI C G+ +Y L +R+ + GT + A+ R+E+L PFP D +++ Y ++R
Sbjct 806 LIFCSGKHFYALLKQRESL----GTKKH---DFAIIRLEELCPFPLDALQQEMSKYKHVR 858
Query 62 TVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNR-IERP 99
V+W+QEEP N G WS+ S R E + L C R + RP
Sbjct 859 DVIWSQEEPQNMGPWSFVSPRFE---KQLACRLRLVSRP 894
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 39/101 (38%), Positives = 54/101 (53%), Gaps = 13/101 (12%)
Query 1 RLILCCGQIYYDLHAERDRI-RSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPN 59
R++ C G+ YY L R+ I +E T AL RVE+L PFP + ++L Y N
Sbjct 810 RVLFCSGKHYYALLKHRETIPEAEKNT--------ALVRVEELCPFPTEALQQELNKYTN 861
Query 60 LRTVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNR-IERP 99
+ +W+QEEP N G WS+ S R E + L C R + RP
Sbjct 862 AKEFIWSQEEPQNMGCWSFVSPRFE---KQLACKLRLVSRP 899
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 50/87 (57%), Gaps = 9/87 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
LI C G+++YDL ER S +VA++R+EQLSPFPFDL + + YP+
Sbjct 900 HLIFCTGKVFYDLQRERK--------SRGLEERVAISRIEQLSPFPFDLVKAEAEKYPHA 951
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCL 87
++W QEE NQG + Y RI + L
Sbjct 952 H-LLWCQEEHKNQGYYDYVKPRISTTL 977
> mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 component
DHKTD1, mitochondrial-like
Length=532
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 37/99 (37%), Positives = 56/99 (56%), Gaps = 11/99 (11%)
Query 2 LILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNLR 61
LI C G+ +Y L +R+ + GT + A+ R+E+L PFP D +++ Y ++R
Sbjct 417 LIFCSGKHFYALLKQRESL----GTKKH---DFAIIRLEELCPFPLDALQQEMSKYKHVR 469
Query 62 TVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNR-IERP 99
V+W+QEEP N G WS+ S R E + L C R + RP
Sbjct 470 DVIWSQEEPQNMGPWSFVSPRFE---KQLACRLRLVSRP 505
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 31/76 (40%), Positives = 46/76 (60%), Gaps = 9/76 (11%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R+++C G++YYDL +R + N + VA+ R+EQL PFP LQ + ++
Sbjct 816 RVVMCSGKVYYDLLEQRRK---------NNQHDVAIVRIEQLYPFPHKAMQEVLQQFAHV 866
Query 61 RTVVWAQEEPMNQGAW 76
+ VW QEEP+NQGAW
Sbjct 867 KDFVWCQEEPLNQGAW 882
> dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1008
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 32/82 (39%), Positives = 48/82 (58%), Gaps = 9/82 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R+I C G++YY+L ER +++ E VA+ R+EQ+SPFPFDL +++ Y N
Sbjct 885 RVIFCTGKVYYELAKERKQLKLE--------ENVAIVRLEQISPFPFDLIKAEVEKYSNA 936
Query 61 RTVVWAQEEPMNQGAWSYTSKR 82
++W QEE N G + Y R
Sbjct 937 E-LIWCQEEHKNMGYYDYIRPR 957
> hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogenase
E1 and transketolase domain containing 1 (EC:1.2.4.2)
Length=919
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 55/99 (55%), Gaps = 11/99 (11%)
Query 2 LILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNLR 61
L+ C G+ +Y L +R+ + ++ + A+ RVE+L PFP D +++ Y +++
Sbjct 805 LVFCSGKHFYSLVKQRESLGAK-------KHDFAIIRVEELCPFPLDSLQQEMSKYKHVK 857
Query 62 TVVWAQEEPMNQGAWSYTSKRIESCLRHLGCPNR-IERP 99
+W+QEEP N G WS+ S R E + L C R + RP
Sbjct 858 DHIWSQEEPQNMGPWSFVSPRFE---KQLACKLRLVGRP 893
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 49/89 (55%), Gaps = 9/89 (10%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
R+I C G++YY+L ER R ++VA+ R+EQ+SPFPFDL ++ + Y
Sbjct 894 RVIFCTGKVYYELVKERHR--------KGLDSQVAITRLEQISPFPFDLVKQEAEKYAT- 944
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCLRH 89
+VW QEE N G + Y R + L H
Sbjct 945 SELVWCQEEHKNMGYYDYVKARFLTILNH 973
> cel:ZK836.2 hypothetical protein
Length=911
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 33/87 (37%), Positives = 46/87 (52%), Gaps = 8/87 (9%)
Query 1 RLILCCGQIYYDLHAERDRIRSEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNL 60
++I G+ + ++ RD E G D+ VA+ RVE L PFP L+ YP
Sbjct 797 KVIFVSGKHWINVEKARD----ERGLKDS----VAIVRVEMLCPFPVVDLQAVLKKYPGA 848
Query 61 RTVVWAQEEPMNQGAWSYTSKRIESCL 87
+ VW+QEEP N GAWS+ R E+ L
Sbjct 849 QDFVWSQEEPRNAGAWSFVRPRFENAL 875
> dre:793708 calcium channel, voltage-dependent, L type, alpha
1D subunit-like; K04853 voltage-dependent calcium channel L
type alpha-1F
Length=2041
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query 26 TSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPN 59
T NP ALA VE PFPFD+ I L I+ N
Sbjct 54 TLSNPIRMAALALVEWKYPFPFDIFIL-LAIFAN 86
> pfa:PF14_0088 aldo/keto reductase, putative
Length=1212
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 10 YYDLHAERDRIRSEYGTSDNPGN 32
Y DL E DRI+S +G +D P +
Sbjct 1011 YTDLKEEHDRIKSTFGLNDEPND 1033
> cel:F26D11.9 clec-217; C-type LECtin family member (clec-217)
Length=257
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 73 QGAWSYTSKRIES-CLRHLGCPNRIERP 99
QG W ++ ES CLR +G PN+I+ P
Sbjct 101 QGDWKMFKRQNESVCLRVMGTPNKIDPP 128
> mmu:212483 Fam193b, IRIZIO, Kiaa1931, MGC29235, MGC38018, MGC39032;
family with sequence similarity 193, member B
Length=778
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 46 PFDLAIRDLQIYPNLRTVVWAQEEPMNQGAWSYTSKRIESCLRHLGCP 93
P + +R L PNL V+W + + +SK + SC + L P
Sbjct 537 PENGLVRRLNTVPNLSRVIWVKTPKPGNPSSEESSKEVPSCKQELSEP 584
> hsa:340152 ZC3H12D, C6orf95, FLJ00361, FLJ46041, MCPIP4, TFL,
dJ281H8.1, p34; zinc finger CCCH-type containing 12D
Length=527
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 22 SEYGTSDNPGNKVALARVEQLSPFPFDLAIRDLQIYPNLRTVV 64
S Y T D+ G+ A AR+ S FP D R + +P L +
Sbjct 466 SVYATEDDEGDARARARIALYSVFPRDQVDRVMAAFPELSDLA 508
> dre:100329822 NOL1/NOP2/Sun domain family, member 6-like
Length=417
Score = 27.7 bits (60), Expect = 9.6, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 21 RSEYGTSDNPGNKVALARVEQLSPFP-FDLAIRDLQIYPNLRTVV 64
RSE +SD PG +A+ E L P FD + D NL +VV
Sbjct 179 RSELFSSDKPGRGLAIRMTEPLYQSPSFDGVLTDELFLQNLPSVV 223
> dre:100149557 NOL1/NOP2/Sun domain family, member 6-like
Length=417
Score = 27.7 bits (60), Expect = 9.6, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 21 RSEYGTSDNPGNKVALARVEQLSPFP-FDLAIRDLQIYPNLRTVV 64
RSE +SD PG +A+ E L P FD + D NL +VV
Sbjct 179 RSELFSSDKPGRGLAIRMTEPLYQSPSFDGVLTDELFLQNLPSVV 223
Lambda K H
0.323 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40