bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7652_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
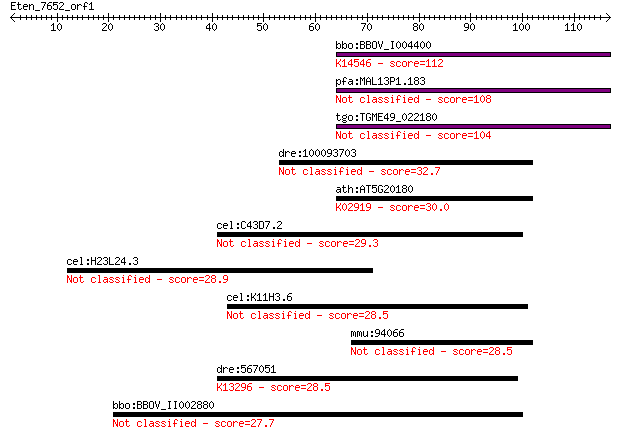
bbo:BBOV_I004400 19.m02216; hypothetical protein; K14546 U3 sm... 112 3e-25
pfa:MAL13P1.183 conserved Plasmodium protein, unknown function 108 6e-24
tgo:TGME49_022180 hypothetical protein 104 7e-23
dre:100093703 mrpl36, si:dkey-261i16.3; mitochondrial ribosoma... 32.7 0.30
ath:AT5G20180 ribosomal protein L36 family protein; K02919 lar... 30.0 2.1
cel:C43D7.2 fbxb-65; F-box B protein family member (fbxb-65) 29.3 3.4
cel:H23L24.3 ttll-11; Tubulin Tyrosine Ligase Like family memb... 28.9 3.9
cel:K11H3.6 hypothetical protein 28.5 5.1
mmu:94066 Mrpl36, AI646041; mitochondrial ribosomal protein L36 28.5 5.7
dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic ... 28.5 6.0
bbo:BBOV_II002880 18.m06237; 85 kDa protein 27.7 9.7
> bbo:BBOV_I004400 19.m02216; hypothetical protein; K14546 U3
small nucleolar RNA-associated protein 5
Length=265
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 46/53 (86%), Positives = 50/53 (94%), Gaps = 0/53 (0%)
Query 64 MWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQALTSPPPRSRGIPKY 116
MWHRG L LLCSSCRYV+RRWHVPILGVDCNANPRHKQAL++P PRSRGIPK+
Sbjct 1 MWHRGTLTLLCSSCRYVIRRWHVPILGVDCNANPRHKQALSNPAPRSRGIPKH 53
> pfa:MAL13P1.183 conserved Plasmodium protein, unknown function
Length=91
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 44/53 (83%), Positives = 50/53 (94%), Gaps = 0/53 (0%)
Query 64 MWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQALTSPPPRSRGIPKY 116
MWHRGVL LLC+SCRYV+ +WHVP+LGVDCNANPRHKQ L++PPPRSRGIPKY
Sbjct 1 MWHRGVLSLLCNSCRYVIHKWHVPLLGVDCNANPRHKQVLSNPPPRSRGIPKY 53
> tgo:TGME49_022180 hypothetical protein
Length=90
Score = 104 bits (259), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 46/53 (86%), Positives = 49/53 (92%), Gaps = 0/53 (0%)
Query 64 MWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQALTSPPPRSRGIPKY 116
MWHRG L LLCSSCRYVVR+WHVPILGVDCNANPRHKQAL+ P PRSRGIPK+
Sbjct 1 MWHRGTLSLLCSSCRYVVRKWHVPILGVDCNANPRHKQALSVPAPRSRGIPKH 53
> dre:100093703 mrpl36, si:dkey-261i16.3; mitochondrial ribosomal
protein L36
Length=116
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 53 QPILSRSPTNLMWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQ 101
Q +L P+ M + L+ C+ C +V RR L V C A+PRHKQ
Sbjct 68 QQLLCVQPSAGMKTKSALKKRCNECFFVRRRGR---LFVFCKAHPRHKQ 113
> ath:AT5G20180 ribosomal protein L36 family protein; K02919 large
subunit ribosomal protein L36
Length=103
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 3/38 (7%)
Query 64 MWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQ 101
M R ++ +C C+ V RR V ++ C++NP+HKQ
Sbjct 1 MKVRSSVKKMCEFCKTVKRRGRVYVI---CSSNPKHKQ 35
> cel:C43D7.2 fbxb-65; F-box B protein family member (fbxb-65)
Length=307
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 28/59 (47%), Gaps = 2/59 (3%)
Query 41 SVTMSTTSPASRQPILSRSPTNLMWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPRH 99
+ + +S PILS + L G +LLC +C+Y++ + P+ D N +H
Sbjct 146 DLDLIVSSSTWYHPILSSNRKYLTLTAGFTDLLCVNCKYLIVQ--QPVTARDLNLMLKH 202
> cel:H23L24.3 ttll-11; Tubulin Tyrosine Ligase Like family member
(ttll-11)
Length=707
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 35/78 (44%), Gaps = 19/78 (24%)
Query 12 RSTAMGSSQENLAPAPASNSEAVQEACSAS--VTMSTTSPASRQPILS------------ 57
RS+++ S EN P+ SNS ++ +AS T+ T+ S Q ++S
Sbjct 93 RSSSLSSIMENRPPSGISNSSFIRSRNTASRRFTIDTSRAKSNQYVVSLCSKKIGIIEYP 152
Query 58 -----RSPTNLMWHRGVL 70
+ P ++ WH VL
Sbjct 153 DGRSDKQPCDVYWHNVVL 170
> cel:K11H3.6 hypothetical protein
Length=77
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 32/65 (49%), Gaps = 13/65 (20%)
Query 43 TMSTTSPASRQPILSRSP----TNLMWHRGVLELLCSSCRYVVR---RWHVPILGVDCNA 95
T+S + RQ + R+P T + L+L C SC Y +R R HV +CN
Sbjct 8 TLSQGNSIIRQLLAVRNPMCQETAGFKVKSRLKLRCRSC-YFLRVEGRLHV-----ECNE 61
Query 96 NPRHK 100
NPRHK
Sbjct 62 NPRHK 66
> mmu:94066 Mrpl36, AI646041; mitochondrial ribosomal protein
L36
Length=102
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 19/35 (54%), Gaps = 3/35 (8%)
Query 67 RGVLELLCSSCRYVVRRWHVPILGVDCNANPRHKQ 101
+GV++ C C V RR IL C NP+HKQ
Sbjct 68 KGVIKKRCKDCYKVKRRGRWFIL---CKTNPKHKQ 99
> dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1117
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 8/58 (13%)
Query 41 SVTMSTTSPASRQPILSRSPTNLMWHRGVLELLCSSCRYVVRRWHVPILGVDCNANPR 98
++T++T PA +PIL+ P + G+ LLC + W+ PI + N R
Sbjct 587 TLTLATLIPAEEKPILAPEP---LLMEGLEPLLCQ-----INNWNFPIFSLVEKTNGR 636
> bbo:BBOV_II002880 18.m06237; 85 kDa protein
Length=574
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 31/79 (39%), Gaps = 5/79 (6%)
Query 21 ENLAPAPASNSEAVQEACSASVTMSTTSPASRQPILSRSPTNLMWHRGVLELLCSSCRYV 80
E A P +E T + +P P+++ TN +WH V V
Sbjct 404 EKPASRPCYGGRKGEEVVVLQTTSTNQTPLKELPVVTGPSTNRLWHENV-----DIDMPV 458
Query 81 VRRWHVPILGVDCNANPRH 99
V++ ILG N +P+H
Sbjct 459 VQKRLSTILGYVTNYDPKH 477
Lambda K H
0.316 0.126 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40