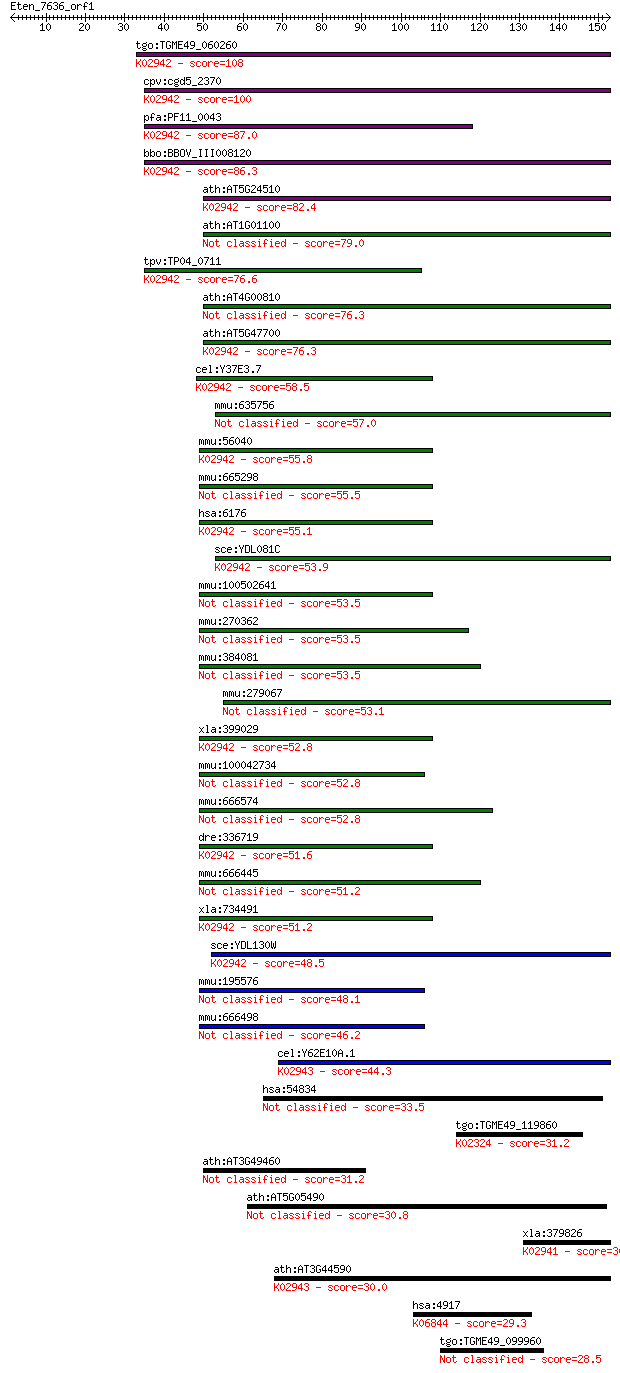
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7636_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_060260 60S acidic ribosomal protein P1, putative ; ... 108 5e-24
cpv:cgd5_2370 60S acidic ribosomal protein LP1 like protein of... 100 1e-21
pfa:PF11_0043 60S ribosomal protein P1, putative; K02942 large... 87.0 2e-17
bbo:BBOV_III008120 17.m07710; 60S acidic ribosomal protein p1;... 86.3 3e-17
ath:AT5G24510 60s acidic ribosomal protein P1, putative; K0294... 82.4 5e-16
ath:AT1G01100 60S acidic ribosomal protein P1 (RPP1A) 79.0 6e-15
tpv:TP04_0711 60S acidic ribosomal protein P1; K02942 large su... 76.6 3e-14
ath:AT4G00810 60S acidic ribosomal protein P1 (RPP1B) 76.3 4e-14
ath:AT5G47700 60S acidic ribosomal protein P1 (RPP1C); K02942 ... 76.3 4e-14
cel:Y37E3.7 rla-1; Ribosomal protein, Large subunit, Acidic (P... 58.5 8e-09
mmu:635756 Gm7166; predicted gene 7166 57.0
mmu:56040 Rplp1, 2410042H16Rik, Arpp1, C430017H15Rik, MGC10313... 55.8 5e-08
mmu:665298 Gm11942, OTTMUSG00000004999; predicted gene 11942 55.5
hsa:6176 RPLP1, FLJ27448, MGC5215, P1, RPP1; ribosomal protein... 55.1 9e-08
sce:YDL081C RPP1A, RPLA1; Ribosomal stalk protein P1 alpha, in... 53.9 2e-07
mmu:100502641 60S acidic ribosomal protein P1-like 53.5 2e-07
mmu:270362 60S acidic ribosomal protein P1-like 53.5 3e-07
mmu:384081 Gm13249, OTTMUSG00000011077; predicted gene 13249 53.5
mmu:279067 Gm13777, OTTMUSG00000014339; predicted gene 13777 53.1
xla:399029 rplp1, MGC68562; ribosomal protein, large, P1; K029... 52.8 4e-07
mmu:100042734 Gm10073; predicted pseudogene 10073 52.8
mmu:666574 Gm13144, OTTMUSG00000010663; predicted gene 13144 52.8
dre:336719 rplp1, MGC77776, fa93f02, wu:fa93f02, zgc:77776; ri... 51.6 1e-06
mmu:666445 Gm13035, OTTMUSG00000010106; predicted gene 13035 51.2
xla:734491 hypothetical protein MGC114621; K02942 large subuni... 51.2 1e-06
sce:YDL130W RPP1B, RPL44', RPLA3; Rpp1bp; K02942 large subunit... 48.5 8e-06
mmu:195576 Gm13229, OTTMUSG00000011032; predicted gene 13229 48.1
mmu:666498 Gm13149, OTTMUSG00000010668; predicted gene 13149 46.2
cel:Y62E10A.1 rla-2; Ribosomal protein, Large subunit, Acidic ... 44.3 2e-04
hsa:54834 GDAP2, FLJ20142, dJ776P7.1; ganglioside induced diff... 33.5 0.29
tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit, pu... 31.2 1.2
ath:AT3G49460 60S acidic ribosomal protein-related 31.2 1.5
ath:AT5G05490 SYN1; SYN1 (SYNAPTIC 1) 30.8 1.8
xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large, P0... 30.0 2.9
ath:AT3G44590 60S acidic ribosomal protein P2 (RPP2D); K02943 ... 30.0 3.3
hsa:4917 NTN3, NTN2L; netrin 3; K06844 netrin 3 29.3
tgo:TGME49_099960 hypothetical protein 28.5 9.0
> tgo:TGME49_060260 60S acidic ribosomal protein P1, putative
; K02942 large subunit ribosomal protein LP1
Length=179
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 70/120 (58%), Positives = 89/120 (74%), Gaps = 0/120 (0%)
Query 33 AKMAAVPTSSLPDGQRQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFA 92
+ MAAV TS+LP+ Q+QELLCTYA+LILSD+ M+++AENI+KLV A+G +VE YMP LFA
Sbjct 60 STMAAVATSALPEAQKQELLCTYAALILSDDKMDVTAENIQKLVVASGNTVEPYMPTLFA 119
Query 93 RALKGHNITDLLAGAGTVAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
RAL+G NI DL++ AG AAA AA A A A +++E EEE+ DMGFSLFD
Sbjct 120 RALQGQNIADLISNAGACAAAAPAAAAPVAGGDAGAAPAKEEKKEEPEEEEDDMGFSLFD 179
> cpv:cgd5_2370 60S acidic ribosomal protein LP1 like protein
of possible plant origin ; K02942 large subunit ribosomal protein
LP1
Length=122
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 62/122 (50%), Positives = 83/122 (68%), Gaps = 4/122 (3%)
Query 35 MAAVPTSSLPDGQRQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARA 94
MAAV + LP Q QEL+C+YA+L+LSD G+ +++ENI+K++SAAG SVE Y PGLFA+A
Sbjct 1 MAAVSMNELPQSQVQELICSYAALVLSDGGVPVTSENIKKIISAAGGSVEPYFPGLFAQA 60
Query 95 LKGHNITDLLAGAGTVAAAPAAAPAAAADNAADKGGKPAKQE----EPEEEEDADMGFSL 150
L N++D++AG G + A A A A A D G A + E EEEE+ D+GFSL
Sbjct 61 LSTTNVSDIVAGCGAASVAVPVAGGAGAGAAQDSGASAAADDKKKKEEEEEEEGDLGFSL 120
Query 151 FD 152
FD
Sbjct 121 FD 122
> pfa:PF11_0043 60S ribosomal protein P1, putative; K02942 large
subunit ribosomal protein LP1
Length=118
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/83 (51%), Positives = 61/83 (73%), Gaps = 1/83 (1%)
Query 35 MAAVPTSSLPDGQRQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARA 94
MA++P S LP+ ++QELLCTYA+LIL +E M I+++NI KL+ + +V Y+P LF RA
Sbjct 1 MASIPASELPECEKQELLCTYAALILHEEKMSITSDNILKLIKNSNNTVLPYLPMLFERA 60
Query 95 LKGHNITDLLAGAGTVAAAPAAA 117
LKG +I LL+ +V +APAAA
Sbjct 61 LKGKDIQSLLSNL-SVGSAPAAA 82
> bbo:BBOV_III008120 17.m07710; 60S acidic ribosomal protein p1;
K02942 large subunit ribosomal protein LP1
Length=118
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 63/118 (53%), Positives = 80/118 (67%), Gaps = 0/118 (0%)
Query 35 MAAVPTSSLPDGQRQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARA 94
M++VP S+L QR ELLC Y+SLIL DEG++ISAENI KL+ A +V+ + P LFA+A
Sbjct 1 MSSVPMSALSAEQRDELLCIYSSLILYDEGLDISAENITKLIKAVDINVQPFRPMLFAKA 60
Query 95 LKGHNITDLLAGAGTVAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
L+G NI +L AG G+ AAA A A A AA+ + K E EEE+ DMGFSLFD
Sbjct 61 LQGKNIAELFAGVGSSAAAAPVAAAGGAPAAAEDKAEAKKPEAEPEEEEDDMGFSLFD 118
> ath:AT5G24510 60s acidic ribosomal protein P1, putative; K02942
large subunit ribosomal protein LP1
Length=111
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 49/108 (45%), Positives = 61/108 (56%), Gaps = 6/108 (5%)
Query 50 ELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLL----- 104
EL CTYA+LIL D+G+EI+AENI KLV A +VESY P LFA+ + NI DL+
Sbjct 5 ELACTYAALILHDDGIEITAENISKLVKTANVNVESYWPSLFAKLCEKKNIDDLIMNVGA 64
Query 105 AGAGTVAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
G G AAP A+ + + K + EE ED DM LFD
Sbjct 65 GGCGVARPVTTAAPTASQSVSIPEEKKNEMEVIKEESED-DMIIGLFD 111
> ath:AT1G01100 60S acidic ribosomal protein P1 (RPP1A)
Length=96
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 45/103 (43%), Positives = 59/103 (57%), Gaps = 12/103 (11%)
Query 50 ELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAGT 109
EL C+YA +IL DEG+ I+A+ I LV AAG S+ESY P LFA+ + N+TDL+ G
Sbjct 6 ELACSYAVMILEDEGIAITADKIATLVKAAGVSIESYWPMLFAKMAEKRNVTDLIMNVGA 65
Query 110 VAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
A A + K++EP EE D D+GF LFD
Sbjct 66 GGGGAAPAA------------EEKKKDEPAEESDGDLGFGLFD 96
> tpv:TP04_0711 60S acidic ribosomal protein P1; K02942 large
subunit ribosomal protein LP1
Length=117
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/70 (48%), Positives = 50/70 (71%), Gaps = 0/70 (0%)
Query 35 MAAVPTSSLPDGQRQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARA 94
M+ VP S L QR+EL+C Y+SL+L D+G++++ +NI KLV AA ++ + P LFARA
Sbjct 1 MSTVPVSELTKEQREELMCVYSSLVLYDDGLDVTQDNILKLVKAAKGDMQPFTPMLFARA 60
Query 95 LKGHNITDLL 104
LKG ++ LL
Sbjct 61 LKGKDLGSLL 70
> ath:AT4G00810 60S acidic ribosomal protein P1 (RPP1B)
Length=113
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 52/108 (48%), Positives = 66/108 (61%), Gaps = 5/108 (4%)
Query 50 ELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA-- 107
EL C+YA +IL DEG+ I+++ I LV AAG +ESY P LFA+ + N+TDL+
Sbjct 6 ELACSYAVMILEDEGIAITSDKIATLVKAAGVEIESYWPMLFAKMAEKRNVTDLIMNVGA 65
Query 108 ---GTVAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
G A AAAPAAA AA K K++EP EE D D+GF LFD
Sbjct 66 GGGGGGAPVSAAAPAAAGGAAAAAPAKEEKKDEPAEESDGDLGFGLFD 113
> ath:AT5G47700 60S acidic ribosomal protein P1 (RPP1C); K02942
large subunit ribosomal protein LP1
Length=113
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 48/108 (44%), Positives = 63/108 (58%), Gaps = 5/108 (4%)
Query 50 ELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAG--- 106
EL C+YA +IL DEG+ I+A+ I LV AAG ++ESY P LFA+ + N+TDL+
Sbjct 6 ELACSYAVMILEDEGIAITADKIATLVKAAGVTIESYWPMLFAKMAEKRNVTDLIMNVGA 65
Query 107 --AGTVAAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
G A AA A AA + K++EP EE D D+GF LFD
Sbjct 66 GGGGGAPVAAAAPAAGGGAAAAAPAAEEKKKDEPAEESDGDLGFGLFD 113
> cel:Y37E3.7 rla-1; Ribosomal protein, Large subunit, Acidic
(P1) family member (rla-1); K02942 large subunit ribosomal protein
LP1
Length=111
Score = 58.5 bits (140), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 48 RQELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
QEL C YA+LIL D+ + I+ E I L+ AA E Y PGLFA+AL+G ++ +L+
Sbjct 4 NQELACVYAALILQDDEVAITGEKIATLLKAANVEFEPYWPGLFAKALEGVDVKNLITSV 63
> mmu:635756 Gm7166; predicted gene 7166
Length=135
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 54/107 (50%), Gaps = 8/107 (7%)
Query 53 CTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAG------ 106
C Y LIL D+ +++ + I L+ A +VES+ PGLF +AL I +
Sbjct 30 CIYFVLILHDQEVKVMKDEINALIKDADFNVESFWPGLFPKALANIKIERFICHLGAGAP 89
Query 107 AGTVAAAPAAAPAAAADNA-ADKGGKPAKQEEPEEEEDADMGFSLFD 152
A PA +P ++ N ++ AK EEP+E D D+GF +FD
Sbjct 90 GSAAGAMPAGSPDSSTTNVPVEERKVEAKNEEPKESVD-DIGFGIFD 135
> mmu:56040 Rplp1, 2410042H16Rik, Arpp1, C430017H15Rik, MGC103133;
ribosomal protein, large, P1; K02942 large subunit ribosomal
protein LP1
Length=114
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + ++ + I L+ AAG SVE + PGLFA+AL NI L+
Sbjct 5 SELACIYSALILHDDEVTVTEDKINALIKAAGVSVEPFWPGLFAKALANVNIGSLICNV 63
> mmu:665298 Gm11942, OTTMUSG00000004999; predicted gene 11942
Length=114
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + ++ + I L+ AAG SVE + PGLFA+AL NI L+
Sbjct 5 SELACIYSALILHDDEVTVTEDKINALIKAAGVSVEPFWPGLFAKALANVNIGCLICNV 63
> hsa:6176 RPLP1, FLJ27448, MGC5215, P1, RPP1; ribosomal protein,
large, P1; K02942 large subunit ribosomal protein LP1
Length=114
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + ++ + I L+ AAG +VE + PGLFA+AL NI L+
Sbjct 5 SELACIYSALILHDDEVTVTEDKINALIKAAGVNVEPFWPGLFAKALANVNIGSLICNV 63
> sce:YDL081C RPP1A, RPLA1; Ribosomal stalk protein P1 alpha,
involved in the interaction between translational elongation
factors and the ribosome; accumulation of P1 in the cytoplasm
is regulated by phosphorylation and interaction with the
P2 stalk component; K02942 large subunit ribosomal protein LP1
Length=106
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/100 (43%), Positives = 58/100 (58%), Gaps = 0/100 (0%)
Query 53 CTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAGTVAA 112
+YA+LIL+D +EIS+E + L +AA VE+ +FA+AL G N+ DLL AA
Sbjct 7 LSYAALILADSEIEISSEKLLTLTNAANVPVENIWADIFAKALDGQNLKDLLVNFSAGAA 66
Query 113 APAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
APA A A + ++EE +EE D DMGF LFD
Sbjct 67 APAGVAGGVAGGEAGEAEAEKEEEEAKEESDDDMGFGLFD 106
> mmu:100502641 60S acidic ribosomal protein P1-like
Length=114
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + ++ + I L+ AAG SVE + PG FA+AL NI L+
Sbjct 5 SELACIYSTLILHDDEVTVTEDKISALIKAAGVSVEPFWPGSFAKALANVNIGSLICNV 63
> mmu:270362 60S acidic ribosomal protein P1-like
Length=111
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAG 108
EL C Y++LIL D + + I L+ AAG S+E + PG FA+AL N L+ G
Sbjct 5 SELACIYSALILEDNEVMVMEVKINALIKAAGVSIEPFWPGSFAKALANVNTGSLICNVG 64
Query 109 TVAAAPAA 116
T AP A
Sbjct 65 TGGPAPLA 72
> mmu:384081 Gm13249, OTTMUSG00000011077; predicted gene 13249
Length=114
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAG 108
EL C Y++LIL D + + I L+ AAG S+E + PG FA+AL N L+ G
Sbjct 5 SELACIYSALILQDNKVMVMDVKINALIKAAGVSIEPFWPGSFAKALANVNTGSLICNVG 64
Query 109 TVAAAPAAAPA 119
PAA A
Sbjct 65 AAEPTPAAGAA 75
> mmu:279067 Gm13777, OTTMUSG00000014339; predicted gene 13777
Length=114
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 57/104 (54%), Gaps = 6/104 (5%)
Query 55 YASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAGTVAAAP 114
Y++LIL D+ + ++ + I L+ +AG SVE + PGLFA+AL NI L+ G P
Sbjct 11 YSALILHDDELTVTEDKINALIKSAGVSVEPFWPGLFAKALANVNIRSLICNVGANGLVP 70
Query 115 AAAPAAAADNAADKGGKPAKQEEPEEEEDA------DMGFSLFD 152
AA A A PA++++ E +++ DMGF LFD
Sbjct 71 AAGAAPAGGPTPSTAAAPAEEKKVEAKKEESEEPEDDMGFGLFD 114
> xla:399029 rplp1, MGC68562; ribosomal protein, large, P1; K02942
large subunit ribosomal protein LP1
Length=113
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + I+ + I L+ AG +VE + P LFA+AL NI L++
Sbjct 5 SELACIYSALILHDDEVSITEDKISTLIKTAGVTVEPFWPSLFAKALANINIGSLISNV 63
> mmu:100042734 Gm10073; predicted pseudogene 10073
Length=114
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLA 105
EL C Y++LIL + + ++ + I L+ AAG SVE + PGLFA+AL NI L+
Sbjct 5 SELACIYSALILHGDEVTVTEDKINALIKAAGVSVEPFWPGLFAKALANVNIGSLIC 61
> mmu:666574 Gm13144, OTTMUSG00000010663; predicted gene 13144
Length=114
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAG 108
+L C Y++LIL D+ + ++ I L+ AAG S+E + PG FA+AL N L+ G
Sbjct 5 SKLTCIYSALILQDKEVMVTEVKINALIKAAGVSIEPFWPGSFAKALANVNTGSLICNVG 64
Query 109 TVAAAPAAAPAAAA 122
P PA AA
Sbjct 65 AAGPTP---PAGAA 75
> dre:336719 rplp1, MGC77776, fa93f02, wu:fa93f02, zgc:77776;
ribosomal protein, large, P1; K02942 large subunit ribosomal
protein LP1
Length=113
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + ++ + + L+ AAG ++E + PGLFA+AL +I L+
Sbjct 5 SELACIYSALILHDDEVTVTEDKLNALIKAAGVTIEPFWPGLFAKALASVDIGSLICNV 63
> mmu:666445 Gm13035, OTTMUSG00000010106; predicted gene 13035
Length=114
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGAG 108
EL C Y++LIL D + ++ I L+ AAG S+E + PG FA+AL N L+
Sbjct 5 SELACIYSALILQDNKVIVTEVKINALIKAAGVSIEPFWPGSFAKALANVNTGSLICNVE 64
Query 109 TVAAAPAAAPA 119
PAA A
Sbjct 65 AAEPTPAAGAA 75
> xla:734491 hypothetical protein MGC114621; K02942 large subunit
ribosomal protein LP1
Length=113
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAGA 107
EL C Y++LIL D+ + I+ + I L+ AG +VE + P LFA+AL +I L++
Sbjct 5 SELACIYSALILHDDEVSITEDKISTLIKTAGVTVEPFWPSLFAKALANIDIGSLISNV 63
> sce:YDL130W RPP1B, RPL44', RPLA3; Rpp1bp; K02942 large subunit
ribosomal protein LP1
Length=106
Score = 48.5 bits (114), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 64/102 (62%), Gaps = 1/102 (0%)
Query 52 LCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLAG-AGTV 110
+ ++A+ IL+D G+EI+++N+ + AAGA+V++ ++A+AL+G ++ ++L+G
Sbjct 5 IISFAAFILADAGLEITSDNLLTITKAAGANVDNVWADVYAKALEGKDLKEILSGFHNAG 64
Query 111 AAAPAAAPAAAADNAADKGGKPAKQEEPEEEEDADMGFSLFD 152
A A A + AA D + K+EE EE D DMGF LFD
Sbjct 65 PVAGAGAASGAAAAGGDAAAEEEKEEEAAEESDDDMGFGLFD 106
> mmu:195576 Gm13229, OTTMUSG00000011032; predicted gene 13229
Length=119
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLA 105
EL C Y++LIL D + ++ I L+ AAG S+E + PGL A+AL N L+
Sbjct 5 SELTCIYSALILQDNEVMVTEVKINALIKAAGVSIEPFWPGLSAKALANVNTGSLIC 61
> mmu:666498 Gm13149, OTTMUSG00000010668; predicted gene 13149
Length=119
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 49 QELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLA 105
EL C Y++LIL + + ++ I L+ AAG S+E + PG FA+AL N L+
Sbjct 5 SELACMYSALILQNNEVMVTEVKINALIKAAGVSIEPFWPGSFAKALANVNTGSLIC 61
> cel:Y62E10A.1 rla-2; Ribosomal protein, Large subunit, Acidic
(P1) family member (rla-2); K02943 large subunit ribosomal
protein LP2
Length=110
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/91 (40%), Positives = 52/91 (57%), Gaps = 7/91 (7%)
Query 69 AENIEKLVSAAGASVESYMPGLFARALKGHNITDLLA--GAGTVAAAPAAAPAAAADNAA 126
++++ ++SA G ++ L L G + +L+A AG V+ + AAPAAAA AA
Sbjct 20 VDDLKNILSAVGVDADAETAKLVVSRLAGKTVEELIAEGSAGLVSVSGGAAPAAAAAPAA 79
Query 127 DK-----GGKPAKQEEPEEEEDADMGFSLFD 152
KPAK+EEP+EE D DMGF LFD
Sbjct 80 GGAAPAADSKPAKKEEPKEESDDDMGFGLFD 110
> hsa:54834 GDAP2, FLJ20142, dJ776P7.1; ganglioside induced differentiation
associated protein 2
Length=496
Score = 33.5 bits (75), Expect = 0.29, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 38/92 (41%), Gaps = 11/92 (11%)
Query 65 MEISAENIEKLVSAAG----ASVESYMPGLFARALKGHN--ITDLLAGAGTVAAAPAAAP 118
+EI E IEK+V A + + +P F R+LK N + L A G P
Sbjct 193 LEIHGETIEKVVFAVSDLEEGTYQKLLPLYFPRSLKEENRSLPYLPADIGNAEGEPVVP- 251
Query 119 AAAADNAADKGGKPAKQEEPEEEEDADMGFSL 150
+ KP E+ +EEED +G L
Sbjct 252 ----ERQIRISEKPGAPEDNQEEEDEGLGVDL 279
> tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit,
putative (EC:2.7.11.12 2.7.7.7); K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=3124
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 114 PAAAPAAAADNAADKGGKPAKQEEPEEEEDAD 145
P P AA + +GG+ AKQ E E+E+D D
Sbjct 2447 PVDGPGVAASESRRQGGRQAKQSEEEDEKDYD 2478
> ath:AT3G49460 60S acidic ribosomal protein-related
Length=46
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 6/41 (14%)
Query 50 ELLCTYASLILSDEGMEISAENIEKLVSAAGASVESYMPGL 90
EL CTYA+L+L D + LV A ++ESY P L
Sbjct 5 ELACTYAALLLHDH------VTMSTLVKTANLNIESYWPSL 39
> ath:AT5G05490 SYN1; SYN1 (SYNAPTIC 1)
Length=617
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 28/116 (24%), Positives = 49/116 (42%), Gaps = 25/116 (21%)
Query 61 SDEGMEISAENIEKLVSAAGASVESYMPGLFA------RALKGHNITDLLAG--AGTVAA 112
S + ++ AE + + + GASVES M G A R N+T +G ++ +
Sbjct 400 SSQNLDSPAEILRTVRTGKGASVESMMAGSRASPETINRQAADINVTPFYSGDDVRSMPS 459
Query 113 APAAAPAAAADN-----------------AADKGGKPAKQEEPEEEEDADMGFSLF 151
P+A AA+ +N + +G +P +E P E + + FS+
Sbjct 460 TPSARGAASINNIEISSKSRMPNRKRPNSSPRRGLEPVAEERPWEHREYEFEFSML 515
> xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large,
P0; K02941 large subunit ribosomal protein LP0
Length=315
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 16/22 (72%), Gaps = 1/22 (4%)
Query 131 KPAKQEEPEEEEDADMGFSLFD 152
K KQEE EE +D DMGF LFD
Sbjct 295 KDEKQEESEESDD-DMGFGLFD 315
> ath:AT3G44590 60S acidic ribosomal protein P2 (RPP2D); K02943
large subunit ribosomal protein LP2
Length=111
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 47/93 (50%), Gaps = 8/93 (8%)
Query 68 SAENIEKLVSAAGASVESYMPGLFARALKGHNITDLLA-GAGTVAAAPAAAPAAAADNAA 126
SA+NI+ ++ A GA V+ L + + G +I +L+A G +A+ P+ A + +
Sbjct 19 SADNIKDIIGAVGADVDGESIELLLKEVSGKDIAELIASGREKLASVPSGGGVAVSAAPS 78
Query 127 DKGGKPAKQEEPEEE-------EDADMGFSLFD 152
GG A E +E D DMGFSLF+
Sbjct 79 SGGGGAAAPAEKKEAKKEEKEESDDDMGFSLFE 111
> hsa:4917 NTN3, NTN2L; netrin 3; K06844 netrin 3
Length=580
Score = 29.3 bits (64), Expect = 5.3, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 103 LLAGAGTVAAAPAAAPAAAADNAADKGGKP 132
LL AGT+ AA + P A AD D+GG P
Sbjct 8 LLLTAGTLFAALSPGPPAPADPCHDEGGAP 37
> tgo:TGME49_099960 hypothetical protein
Length=401
Score = 28.5 bits (62), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 110 VAAAPAAAPAAAADNAADKGGKPAKQ 135
A APA AP A AD+AA G P KQ
Sbjct 2 TADAPAVAPLAEADDAAQASGAPQKQ 27
Lambda K H
0.315 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40