bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7616_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
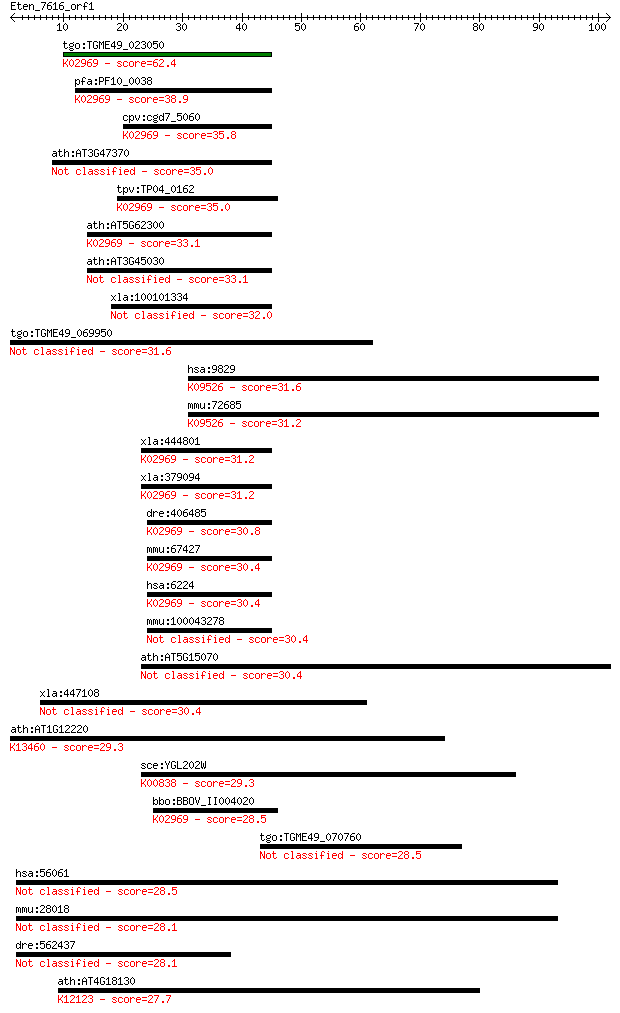
tgo:TGME49_023050 40s ribosomal protein S20, putative ; K02969... 62.4 3e-10
pfa:PF10_0038 40S ribosomal protein S20e, putative; K02969 sma... 38.9 0.004
cpv:cgd7_5060 40S ribosomal protein S20 ; K02969 small subunit... 35.8 0.034
ath:AT3G47370 40S ribosomal protein S20 (RPS20B) 35.0 0.054
tpv:TP04_0162 40S ribosomal protein S20; K02969 small subunit ... 35.0 0.063
ath:AT5G62300 40S ribosomal protein S20 (RPS20C); K02969 small... 33.1 0.22
ath:AT3G45030 40S ribosomal protein S20 (RPS20A) 33.1 0.22
xla:100101334 hypothetical protein LOC100101334 32.0 0.48
tgo:TGME49_069950 hypothetical protein 31.6 0.58
hsa:9829 DNAJC6, DJC6, KIAA0473, MGC129914, MGC129915, MGC4843... 31.6 0.67
mmu:72685 Dnajc6, 2810027M23Rik, mKIAA0473; DnaJ (Hsp40) homol... 31.2 0.80
xla:444801 MGC82136 protein; K02969 small subunit ribosomal pr... 31.2 0.81
xla:379094 rps20, MGC52591; ribosomal protein S20; K02969 smal... 31.2 0.84
dre:406485 rps20, wu:fb81b03, zgc:77758; ribosomal protein S20... 30.8 1.2
mmu:67427 Rps20, 4632426K06Rik, Dsk4, MGC102408; ribosomal pro... 30.4 1.3
hsa:6224 RPS20, FLJ27451, MGC102930; ribosomal protein S20; K0... 30.4 1.3
mmu:100043278 Gm4332; predicted gene 4332 30.4
ath:AT5G15070 acid phosphatase/ oxidoreductase/ transition met... 30.4 1.5
xla:447108 anxa8, MGC85309, anx8; annexin A8 30.4 1.5
ath:AT1G12220 RPS5; RPS5 (RESISTANT TO P. SYRINGAE 5); nucleot... 29.3 3.2
sce:YGL202W ARO8; Aro8p (EC:2.6.1.27 2.6.1.57 2.6.1.5); K00838... 29.3 3.5
bbo:BBOV_II004020 18.m06332; 40S ribosomal protein S20; K02969... 28.5 4.8
tgo:TGME49_070760 hypothetical protein 28.5 5.3
hsa:56061 UBFD1, FLJ38870, FLJ42145, UBPH; ubiquitin family do... 28.5 6.1
mmu:28018 Ubfd1, AI467302, D7Wsu105e, D7Wsu128e, Ubph; ubiquit... 28.1 6.5
dre:562437 ubfd1, MGC162578, im:6905175, zgc:162578; ubiquitin... 28.1 7.5
ath:AT4G18130 PHYE; PHYE (PHYTOCHROME DEFECTIVE E); G-protein ... 27.7 9.2
> tgo:TGME49_023050 40s ribosomal protein S20, putative ; K02969
small subunit ribosomal protein S20e
Length=233
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/35 (88%), Positives = 32/35 (91%), Gaps = 0/35 (0%)
Query 10 ASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
A MSK +K GLEGEEQRLHRIRITLTSKDLKSIER
Sbjct 114 AKMSKLMKGGLEGEEQRLHRIRITLTSKDLKSIER 148
> pfa:PF10_0038 40S ribosomal protein S20e, putative; K02969 small
subunit ribosomal protein S20e
Length=118
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 12 MSKALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
MSK +K ++ E+ RL RIRI LTSK+L++IE+
Sbjct 1 MSKLMKGAIDNEKYRLRRIRIALTSKNLRAIEK 33
> cpv:cgd7_5060 40S ribosomal protein S20 ; K02969 small subunit
ribosomal protein S20e
Length=135
Score = 35.8 bits (81), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 18/27 (66%), Positives = 23/27 (85%), Gaps = 2/27 (7%)
Query 20 LEGEEQR--LHRIRITLTSKDLKSIER 44
GE+Q+ LH+IRITL+SK+LKSIER
Sbjct 24 FNGEQQQAPLHKIRITLSSKNLKSIER 50
> ath:AT3G47370 40S ribosomal protein S20 (RPS20B)
Length=122
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 8 LAASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
+A K K GLE +++H+IRITL+SK++K++E+
Sbjct 1 MAYEPMKPTKAGLEAPLEQIHKIRITLSSKNVKNLEK 37
> tpv:TP04_0162 40S ribosomal protein S20; K02969 small subunit
ribosomal protein S20e
Length=116
Score = 35.0 bits (79), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 19 GLEGEEQRLHRIRITLTSKDLKSIERG 45
G E+ R+H+IR+TL S+DLKSIE+
Sbjct 9 GEYDEDNRIHKIRLTLVSRDLKSIEKA 35
> ath:AT5G62300 40S ribosomal protein S20 (RPS20C); K02969 small
subunit ribosomal protein S20e
Length=124
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
K K GLE +++H+IRITL+SK++K++E+
Sbjct 9 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEK 39
> ath:AT3G45030 40S ribosomal protein S20 (RPS20A)
Length=124
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
K K GLE +++H+IRITL+SK++K++E+
Sbjct 9 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEK 39
> xla:100101334 hypothetical protein LOC100101334
Length=120
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 18 EGLEGEEQRLHRIRITLTSKDLKSIER 44
+ L ++Q++HRIRITLTS ++K++E+
Sbjct 8 KALAEDDQQIHRIRITLTSLNVKNLEK 34
> tgo:TGME49_069950 hypothetical protein
Length=944
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 34/62 (54%), Gaps = 7/62 (11%)
Query 1 FIHPFSSLAASMSKALKEGLEGEE-QRLHRIRITLTSKDLKSIERGEQQRKNDISAFLEV 59
F+ P+ SL ++ KE L EE + LHR +D ++ RGE +R+ D+S +E
Sbjct 554 FLGPYDSLTVFATQKYKELLRQEEHEHLHR-----QDRDTPNL-RGESRRETDVSFMIEA 607
Query 60 LK 61
L+
Sbjct 608 LE 609
> hsa:9829 DNAJC6, DJC6, KIAA0473, MGC129914, MGC129915, MGC48436;
DnaJ (Hsp40) homolog, subfamily C, member 6 (EC:3.1.3.48);
K09526 DnaJ homolog subfamily C member 6
Length=913
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 40/80 (50%), Gaps = 17/80 (21%)
Query 31 RITLTSKDLKSIERGEQQRKNDISAFLEVLKPFCSRYM-----------ALQTRNRHMRL 79
RI + S L +++ G + + +DI +FL+ SR++ + +T H R+
Sbjct 72 RIIVMSFPLDNVDIGFRNQVDDIRSFLD------SRHLDHYTVYNLSPKSYRTAKFHSRV 125
Query 80 SKTLKPISQWPSLYKLWRHC 99
S+ PI Q PSL+ L+ C
Sbjct 126 SECSWPIRQAPSLHNLFAVC 145
> mmu:72685 Dnajc6, 2810027M23Rik, mKIAA0473; DnaJ (Hsp40) homolog,
subfamily C, member 6 (EC:3.1.3.48); K09526 DnaJ homolog
subfamily C member 6
Length=968
Score = 31.2 bits (69), Expect = 0.80, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 40/80 (50%), Gaps = 17/80 (21%)
Query 31 RITLTSKDLKSIERGEQQRKNDISAFLEVLKPFCSRYM-----------ALQTRNRHMRL 79
RI + S + S++ G + + +DI +FL+ SR++ + +T H R+
Sbjct 127 RIIVMSFPVDSVDIGFRNQVDDIRSFLD------SRHLDHYTVYNLSPKSYRTAKFHSRV 180
Query 80 SKTLKPISQWPSLYKLWRHC 99
S+ PI Q PSL+ L+ C
Sbjct 181 SECSWPIRQAPSLHNLFAVC 200
> xla:444801 MGC82136 protein; K02969 small subunit ribosomal
protein S20e
Length=119
Score = 31.2 bits (69), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 20/22 (90%), Gaps = 0/22 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIER 44
+E +HRIRITLTS+++KS+E+
Sbjct 13 QEVAIHRIRITLTSRNVKSLEK 34
> xla:379094 rps20, MGC52591; ribosomal protein S20; K02969 small
subunit ribosomal protein S20e
Length=119
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 20/22 (90%), Gaps = 0/22 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIER 44
+E +HRIRITLTS+++KS+E+
Sbjct 13 QEVAIHRIRITLTSRNVKSLEK 34
> dre:406485 rps20, wu:fb81b03, zgc:77758; ribosomal protein S20;
K02969 small subunit ribosomal protein S20e
Length=119
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +HRIRITLTS+++KS+E+
Sbjct 14 EVAIHRIRITLTSRNVKSLEK 34
> mmu:67427 Rps20, 4632426K06Rik, Dsk4, MGC102408; ribosomal protein
S20; K02969 small subunit ribosomal protein S20e
Length=119
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +HRIRITLTS+++KS+E+
Sbjct 14 EVAIHRIRITLTSRNVKSLEK 34
> hsa:6224 RPS20, FLJ27451, MGC102930; ribosomal protein S20;
K02969 small subunit ribosomal protein S20e
Length=119
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +HRIRITLTS+++KS+E+
Sbjct 14 EVAIHRIRITLTSRNVKSLEK 34
> mmu:100043278 Gm4332; predicted gene 4332
Length=115
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +HRIRITLTS+++KS+E+
Sbjct 10 EVAIHRIRITLTSRNVKSLEK 30
> ath:AT5G15070 acid phosphatase/ oxidoreductase/ transition metal
ion binding
Length=1049
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 23 EEQRLHRIRITLTSKDLKSIERGEQQRKNDISAFLEVLKPFCSRYMALQTRNRHMRLSKT 82
+E R + ++ + S +L+ GE + ++D L P +Y + T RH+R
Sbjct 822 KEDRYSQPKLFVKSDELRRPSTGENKEEDDDKETKYRLDP---KYANVMTPERHVRTRLY 878
Query 83 LKPISQWPSLYKLWRHCNI 101
S SL + R+CN+
Sbjct 879 FTSESHIHSLMNVLRYCNL 897
> xla:447108 anxa8, MGC85309, anx8; annexin A8
Length=363
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 6 SSLAASMSKALKEGLEGEEQRLHRIRITLTSKDLKSI-ERGEQQRKNDISAFLEVL 60
S + KAL L+GE + L +D K++ E GE+ +K D+S F+E+
Sbjct 187 SDTSGDFQKALLILLKGERNEDCYVNEDLAERDAKALYEAGEKNKKADVSVFIEIF 242
> ath:AT1G12220 RPS5; RPS5 (RESISTANT TO P. SYRINGAE 5); nucleotide
binding; K13460 disease resistance protein RPS5
Length=889
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 39/88 (44%), Gaps = 15/88 (17%)
Query 1 FIHPFSSLAASMSKALK---------------EGLEGEEQRLHRIRITLTSKDLKSIERG 45
+IH S AS+ KA++ E G +QRL ++++ LTS + +
Sbjct 28 YIHNLSKNLASLQKAMRMLKARQYDVIRRLETEEFTGRQQRLSQVQVWLTSVLIIQNQFN 87
Query 46 EQQRKNDISAFLEVLKPFCSRYMALQTR 73
+ R N++ L FCS+ + L R
Sbjct 88 DLLRSNEVELQRLCLCGFCSKDLKLSYR 115
> sce:YGL202W ARO8; Aro8p (EC:2.6.1.27 2.6.1.57 2.6.1.5); K00838
aromatic amino acid aminotransferase I [EC:2.6.1.57 2.6.1.27
2.6.1.5]
Length=500
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 6/66 (9%)
Query 23 EEQRLHRIRITLTSKDLKSIERGEQQRKNDISAFLEVLKPFCSRYMALQTRNRHMRL--- 79
E++ + +++ KDLK E+ + K D F LK + +++L T R +R+
Sbjct 247 EDEPYYFLQMNPYIKDLKEREKAQSSPKQDHDEF---LKSLANTFLSLDTEGRVIRMDSF 303
Query 80 SKTLKP 85
SK L P
Sbjct 304 SKVLAP 309
> bbo:BBOV_II004020 18.m06332; 40S ribosomal protein S20; K02969
small subunit ribosomal protein S20e
Length=120
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 25 QRLHRIRITLTSKDLKSIERG 45
R+H+I +TLTS +LK+IE+
Sbjct 19 NRIHKIHVTLTSMNLKAIEKA 39
> tgo:TGME49_070760 hypothetical protein
Length=1615
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 43 ERGEQQRKNDISAFLEVLKPFCSRYMALQTRNRH 76
ER +Q R++ AF VL P CS L+TR H
Sbjct 583 ERQKQHREHRSEAFTFVLPPGCSITTPLKTRTVH 616
> hsa:56061 UBFD1, FLJ38870, FLJ42145, UBPH; ubiquitin family
domain containing 1
Length=309
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 22/100 (22%), Positives = 42/100 (42%), Gaps = 16/100 (16%)
Query 2 IHPFSSLAASMSKALKEGLEGEEQRLHRIRITLTSK---------DLKSIERGEQQRKND 52
IH + L +M K + +GL E++ L I++T +K D+ ++ + + D
Sbjct 114 IHSITGLPPAMQKVMYKGLVPEDKTLREIKVTSGAKIMVVGSTINDVLAVNTPKDAAQQD 173
Query 53 ISAFLEVLKPFCSRYMALQTRNRHMRLSKTLKPISQWPSL 92
A +P C + +H ++ KP PS+
Sbjct 174 AKAEENKKEPLCR-------QKQHRKVLDKGKPEDVMPSV 206
> mmu:28018 Ubfd1, AI467302, D7Wsu105e, D7Wsu128e, Ubph; ubiquitin
family domain containing 1
Length=368
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 22/100 (22%), Positives = 42/100 (42%), Gaps = 16/100 (16%)
Query 2 IHPFSSLAASMSKALKEGLEGEEQRLHRIRITLTSK---------DLKSIERGEQQRKND 52
IH + L +M K + +GL E++ L I++T +K D+ ++ + + D
Sbjct 173 IHSITGLPPAMQKVMYKGLVPEDKTLREIKVTSGAKIMVVGSTINDVLAVNTPKDAAQQD 232
Query 53 ISAFLEVLKPFCSRYMALQTRNRHMRLSKTLKPISQWPSL 92
A +P C + +H ++ KP PS+
Sbjct 233 AKAEENKKEPLCR-------QKQHRKVLDKGKPEDVMPSV 265
> dre:562437 ubfd1, MGC162578, im:6905175, zgc:162578; ubiquitin
family domain containing 1
Length=279
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 2 IHPFSSLAASMSKALKEGLEGEEQRLHRIRITLTSK 37
IH + L +M K + +GL EE+ L I++T +K
Sbjct 84 IHALTGLPPAMQKVMYKGLLPEEKTLREIKVTNGAK 119
> ath:AT4G18130 PHYE; PHYE (PHYTOCHROME DEFECTIVE E); G-protein
coupled photoreceptor/ protein histidine kinase/ signal transducer;
K12123 phytochrome E
Length=1112
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 9 AASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIERGEQQRKNDISAFLEVLKPFCSRYM 68
++ +S + ++ LE + +I + S DLKSIE G+ Q + + +L S+ M
Sbjct 898 SSEISASQRQFLETSDACEKQITTIIESTDLKSIEEGKLQLETEEFRLENILDTIISQVM 957
Query 69 -ALQTRNRHMRL 79
L+ RN +R+
Sbjct 958 IILRERNSQLRV 969
Lambda K H
0.322 0.133 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40